# 1: Life Science x AI
Welcome back to your weekly dose of AI news for Life Science!
Here’s what we have for you this week:
New open source state-of-the-art LLM for medical applications 👀
Good week for open-source code companion … and why do we care 🖥️
Life Science tools of the week 🛠️
Don’t forget to take our short survey to help us understand how you currently use AI in your day-to-day!
New open source state-of-the-art LLM for medical applications 👀
ChatGPT, powered by GPT-3.5, has sparked a revolution in the world of AI-driven conversational tools. Its inception marked a significant advancement in natural language processing, paving the way for the development of various innovative applications. Now the medical industry is embracing the potential of large language models (LLMs) like ChatGPT to enhance patient care, research, and diagnostics.
Recently, researchers have released OpenBioLLM, a fully open-source state-of-the-art tool tailored for biomedical applications. But … what can this tool do?
Here is a list of use cases
Summarise clinical notes, such as EHR data, discharge summaries
Answer medical questions (obviously the information provided should be double-check with a professional)
Extract key insights from notes, such as diseases, symptoms, medications, procedures, and anatomical structures
De-identification by detecting and removing personally identifiable information (PII) from medical records, ensuring patient privacy and compliance with data protection regulations like HIPAA
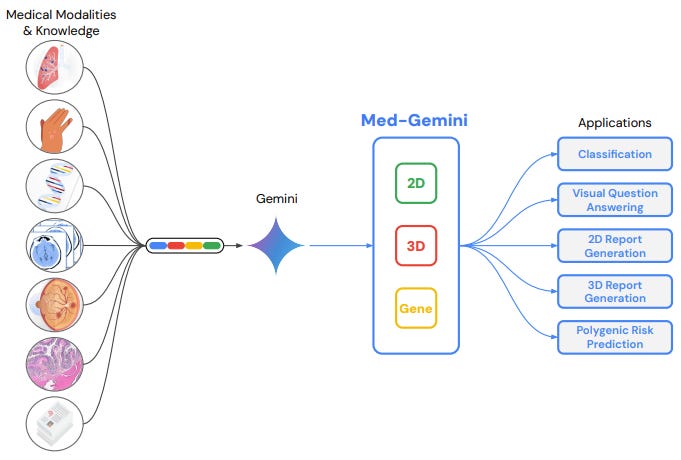
Google is also investing a lot of effort into medical LLMs. They recently published an article on their latest family of Gemini tools for medicine (called Med-Gemini). These family of tools will be able to help doctors to suggest diagnosis, classify and understand radiology images and run Polygenic Risk Score to identify people at high genetic risk for diseases such as pneumonia, stroke, major depression!
If you don’t have time to read new papers, make sure to bookmark the Open Medical-LMM Leaderboard which contains the best LLM models for medical applications, including how they score in specific tasks!
Good week for open-source code companion … and why do we care 🖥️
This has been a good week for open-source code companion
Why are code companion important? Imagine you want to get a plot, interrogate data, run some analysis or develop your own tools. All these tasks require writing code. Problem is that writing code is difficult and time consuming. Instead, you can use a code companion to transform your question/request into code for you! Having open-source code companion allow you to fine-tune the model (ie. tailor them) to your own use cases, making them even more useful!
Three important tools have been released and improved.
1/ StarCoder2-Instruct
StarCoder2 has been gained a lot of traction in the past months as one of the best open source code companion, especially for uncommon languages like Perl. The limitation? You could not chat with it, it mostly acing as a code auto-completion. That has changed with StarCoder2-Instruct! Now you can give the tool instruction via a chat and StarCoder will write the code for you! If you want to run it on your computer, check out Ollama
2/ DeepSeek V2
Deepseek is another well recognised open source code companion. It is specialised on more general languages (like Python) as opposed to StarCoder2 which has been trained on many uncommon languages (eg. Perl). What set DeepSeek v2 aside is the shire size of the model and the amount of data it was trained on. What does this mean in practice? It can generate a lot of good code fast and it is approaching the latest version of GPT4! Also, you can use DeepSeek directly in your browser.
3/ IBM releases Granite models
IBM has entered the game of open-source code companion and has released their Granite code companion to help users to generate, fix and explain their code. The models have been trained on a massive dataset of 500 million lines of code in over 50 programming languages!
Life Science tools of the week 🛠️
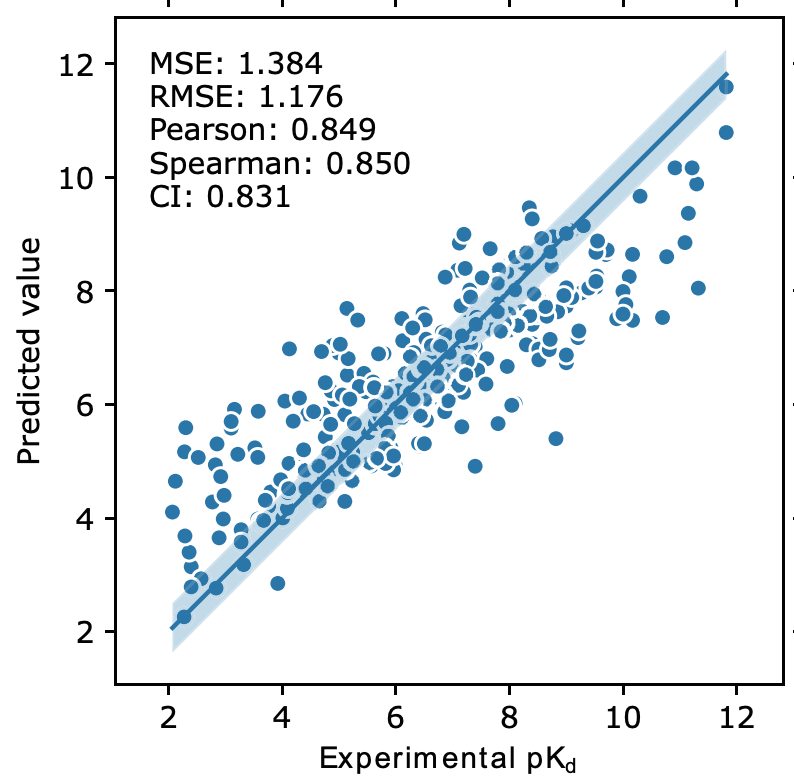
1/ DeepRLI - Protein design
DeepRLI is a a new framework for universal protein–ligand interaction prediction. How is it different than other methods? It implements a multi-objective learning strategy that includes scoring, docking, and screening as optimization goal. The authors report high correlation (> 0.8) between the predicted pKd and the experimental one. For everyone who does not know, the pKd is a measure of the binding affinity of a ligand (such as a drug) to a receptor. While the high correlation does not tell the full story of the protein design … the results are certainly encouraging!
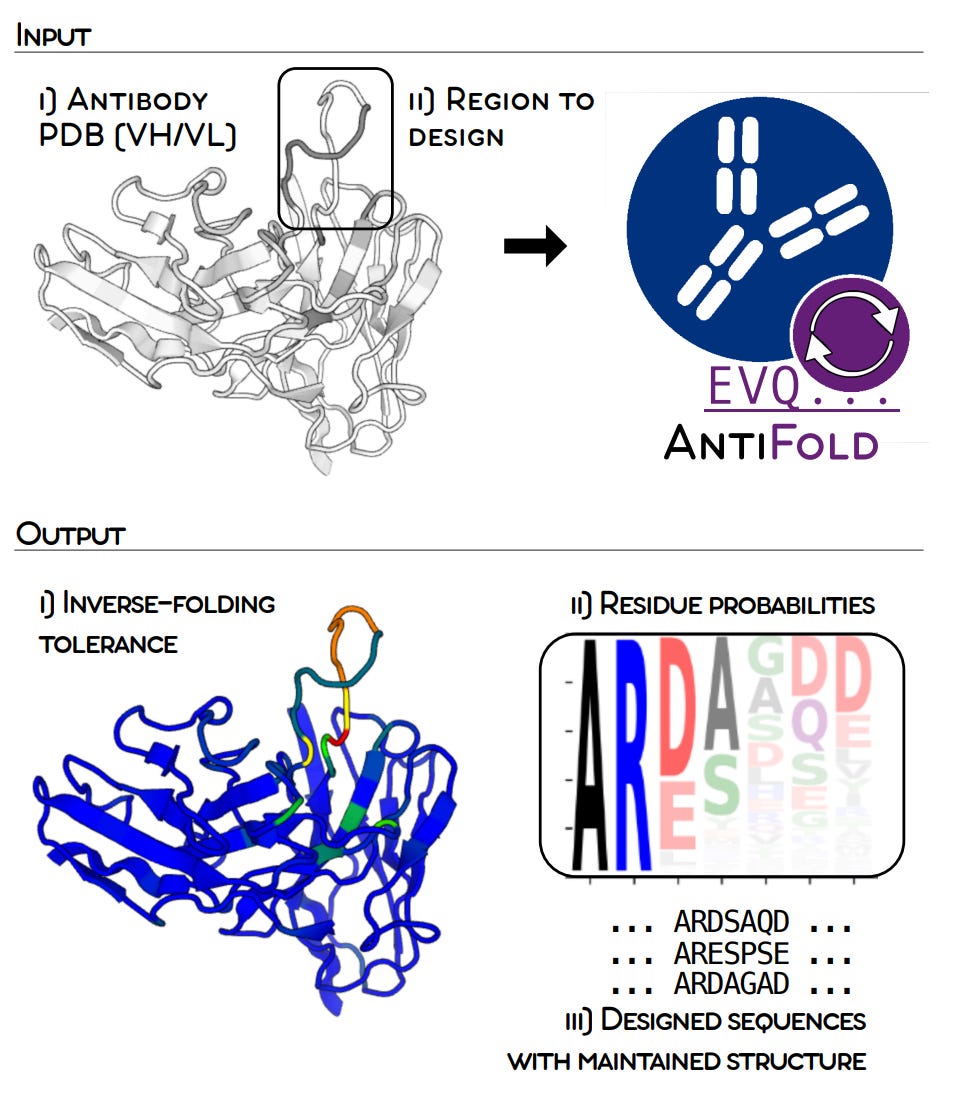
2/ AntiFold - antibody design
The bad news: The design and optimization of antibodies is a tedious process that requires an intricate balance across multiple properties.
The good news: AntiFold is here to help! AntiFold is a fine-tuned version of ESM-IF1 (ie. a version of ESM-IF1 trained for specific use cases) that achieves state-of-the-art in optimising antibodies by leveraging inverse folding. It improves on existing tools on variety fo tasks, such as Antibody-Antigen binding affinity, Sequence design and amino acid recovery.
You can check out a web version of the tool in here
3/ OpenCRISPR - CRISPR-Cas system design
The CRISPR-Cas system is an amazing programmable gene editing system important for gene therapy and personalised medicine. Put it simply, it allows us to edit, modify and replace a specific gene in a specific region. However, CRISPR-based gene editors derived from microbes, while powerful, often show significant functional tradeoffs when ported into non-native environments, such as human cells. Here comes OpenCRISPR, a new AI tool to generate new CRISPR-Cas families and optimise them. The authors of the paper managed to: 1) Generate 4.8x more CRISPR-Cas proteins and 2) Show that ~10% of the newly generated proteins have better activity and specificity compared to the reference SpCas9 … while having 400 different amino acids!
You can use OpenCRISPR tool for free for commercial and academic usage … which means from today onwards you can design your own CRISPR system!
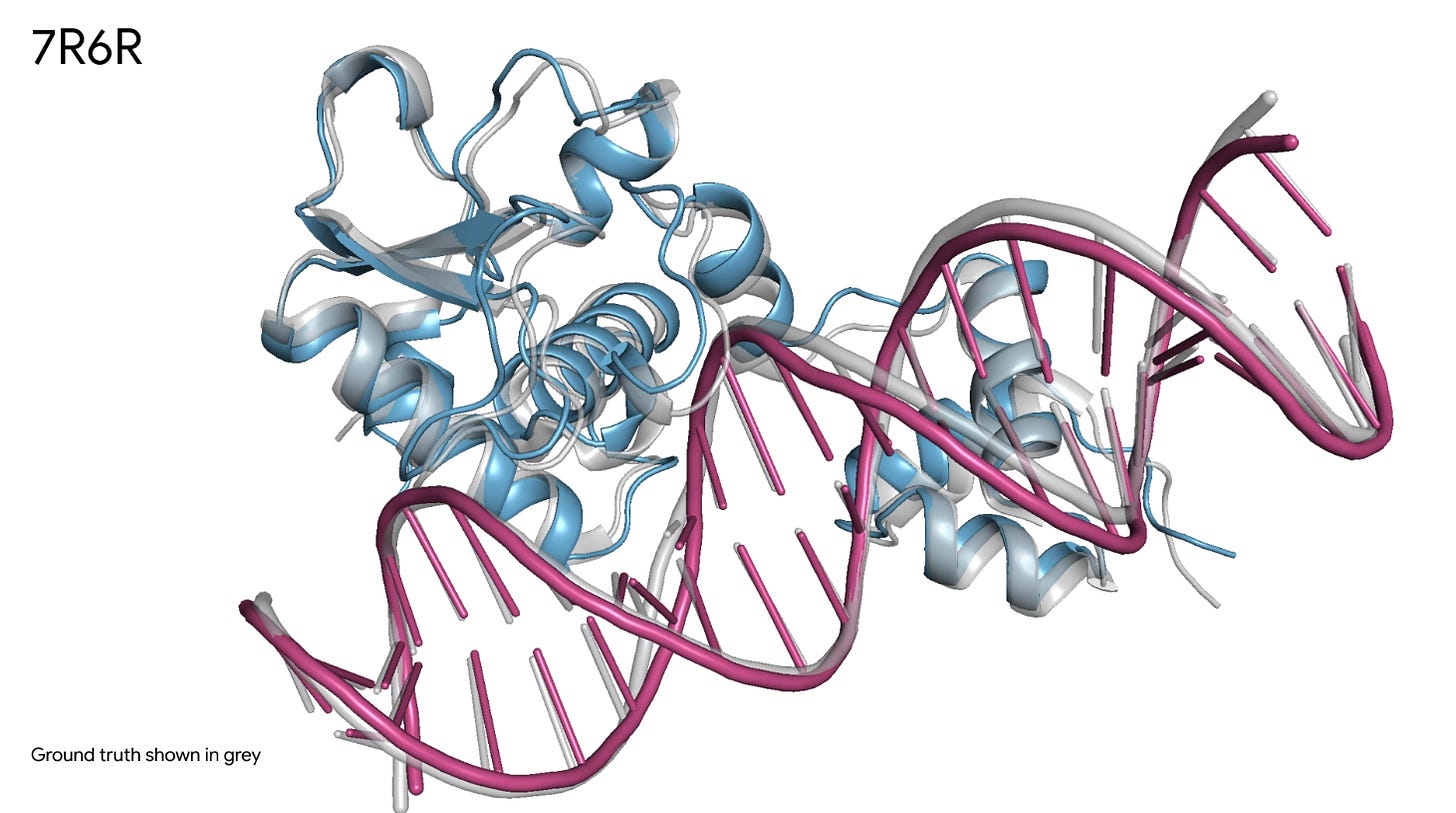
4/ AlphaFold 3 - Protein design
For those of you who don’t know it, AlphaFold is a cornerstone tool to predict protein folding based on their sequence. On May 8th Isomorphic lab, a spinoff of DeepMind, released their third version of the software. You might wonder what’s new. Well, first, the new version expands beyond proteins and now includes folding and interaction between protein, DNA, RNA and small molecules. Second, it provides 50% better prediction compared to the best traditional methods out there!
If you want to try AlphaFold 3 by yourself you can check out the AlphaFold Server. Don’t forget to check out their demo!
BITE-SIZED COOKIES FOR THE WEEK 🍪
You heard a lot about AI in Life Science but don’t know where is heading? Check out the latest report from McKinsey where they explain the current state of the art and the different use cases where AI can help you and your organisation!
There is a lot of hype about AI, but has it delivered in terms of drugs? A recent paper from Boston Consulting Group analysed clinical trials and found our that AI-discovered molecules are 20-30% more likely to pass Phase I clinical trial compared to traditional drugs
How can you increase the bio-security AND IP-protection of your newly designed protein? With ProteinWatermark! This tool integrates with state-of-the-art AI protein design tools like ProteinMPNN & ProGen2 while adding “watermarks” to your protein!
Struggle to easily create images of macromolecular structures? No more! With PDBImages you can simply provide a protein entry (form PDB, AlphaFold2 or custom structures) and it will generate 9 different types of imaging! And the best part … it is open source! Check out the tool in here
If you are looking for more to protein design tools check out POLYGON! It leverages polypharmacology information and deep generative chemistry to generate new compounds! The authors derived and tested 32 compounds targeting MEK1 and mTOR and showed >50% reduction in each protein activity and in cell viability when dosed at 1–10 μM. You can check out the tool yourself in here
Don’t forget to take our short survey to help us understand how you currently use AI in your day-to-day!