BioCypher & BioChatter: Knowled Graph Builder and LLM
Welcome to the “Deep Dive“ series where we bring to you some cool AI tools that have a strong impact in Life Science. In each episode, we bring an exclusive interview with the main developers behind cool tools and we explain to you why the tool is important, what it does and what the future holds!
Today we present BioCypher/BioChatter, two integrated tools to streamline and democratise access to biological insights. We have interviewed the project lead, Sebastian Lobentanzer, a Postdoctoral Researcher at the University Hospital Heidelberg, who shed some light on why BioCypher/BioChatter were created!
🔴 The Problem
There is a lot of Life Science data out there, with new data generated every day. How can researchers properly leverage this information to advance science, when the different data sources are fragmented and not interoperable? One popular approach is a Knowledge Graph (KG). A KG is like a giant map that connects different pieces of biological information, such as genes, proteins, diseases, drugs, and other biological entities, and shows how they relate to each other. This helps researchers understand complex biological systems, discover new relationships, and find patterns that can lead to medical breakthroughs. In a KG, data is represented as the nodes of the graph while the relationship between data is shown as the edges (or link) between the nodes. See below an example on proteins based on Gene Ontology.
Now that we have established that KGs are useful, are KGs for Life Science easily available? Yes BUT they are often not maintained, flexible or adhere to FAIR (Findable, Accessible, Interoperable and Reusable) principles. The main problem stems from the complexity of building KGs, where KGs are built differently depending on the applications and data in mind. For example, a KG for drug discovery (genes are represented as protein ancestors) has a different approach than a KG for a molecular tumour board (genes are represented as clinical markers). This complexity led to the development of many bespoke KGs which do not generalise across different use cases (Lobentanzer et al., 2023).
💡The Idea
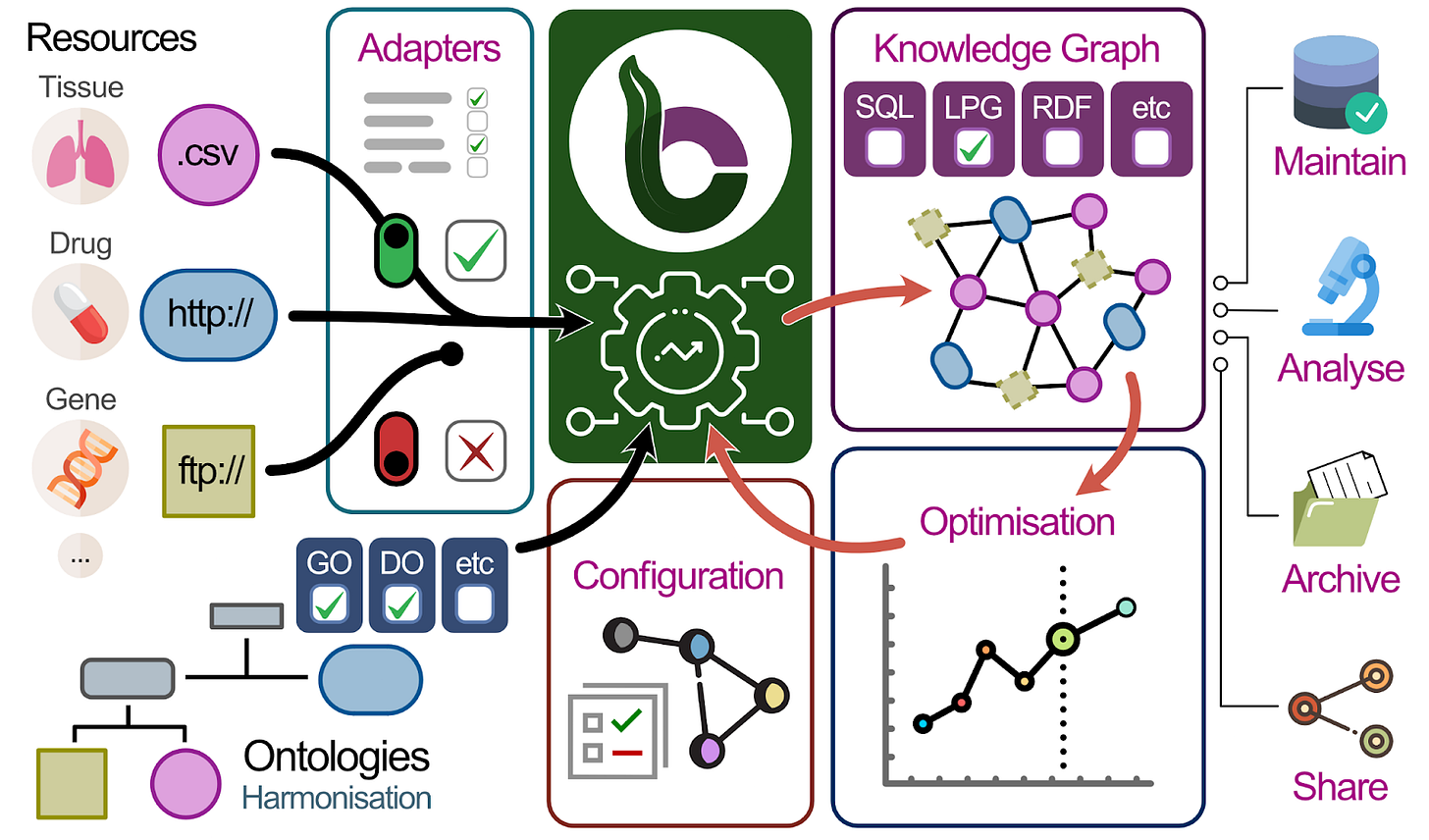
That’s when Sebastian and the team at Julio Saez-Rodriguez’s lab started to think about BioCypher! The idea is to have a single graph which incorporates many different data sources and ontologies in a flexible and scalable way. The users have a simple configuration file where they can define the data structures and their properties to customise their KG. Since it’s fully open-source, every researcher can deploy their own local version (with their own local data).
The beauty of BioCypher is in its modularity, where Sebastian and team are creating “adapters” to connect specific biological data sources and ontologies (GTEx, OpenTargets, UniProt, IntAct, full list in here) to the KG. More importantly, this endeavour is not limited to insiders; as a community project, the creation of adapters and pipelines is a community effort. Several BioCypher-driven knowledge graphs already exist in areas of genetics, oncology, drug discovery, and clinical research.
Now that a KG is created, how can researchers use it? Traditionally, users had to know the exact schema and contents of the graph and learn to use a query language such as SQL or Cypher or had to visualise and filter the KG from a UI. However, this becomes a daunting task when you have a large amount of data and relationships, and many researchers don’t have the background or bandwidth to become database engineers.
That’s where BioChatter comes into play! BioChatter provides a chat-like experience (similar to ChatGPT) for users to query their local BioCypher KG as well as user documents. Imagine using ChatGPT with information on the relationship between genes, proteins, pathways and more!
For those who want more technical details, BioChatter works as a multi-agent RAG system on the KG and/or a vector database for RAG and can use GPT or open-source LLMs (e.g., Llama3). If you want to learn more details, check out the official documentation!
🔮The Future
What does the future hold for BioCypher and BioChatter? A lot! Sebastian and team will keep on maintaining and expanding the features, especially since they have grants to cover the next years of development. Here a list of things they are planning to do in the next 1-2 years:
Expand the BioChatter benchmark on open-source LLMs to understand how these models perform for different tasks.
Add more adaptors such as ChEMBL and Cell Ontology (see here for the ones already planned)
Develop a new module called “BioGather” to extract insights from text and images from resources like papers
Work together with Open Targets to integrate some LLM features to more easily discover data within the Open Targets platform
And the best part … all these tools are open-source from day 1!
If you want to get involved, contact Sebastian!