AI in Life Science: Weekly Insights
Weekly Insights | November 14, 2024
In this issue:
Welcome back to your weekly dose of AI news for Life Science!
This week, we have some exciting new models lined up for you:
Dive into these game-changing innovations and explore how they are transforming the biotech and healthcare landscapes!
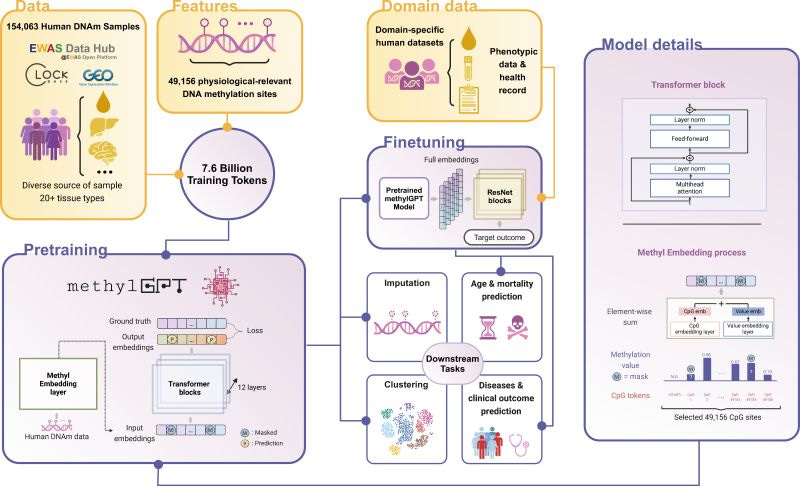
MethylGPT: New foundation model for DNA methylation 🧬
Another week and another DNA methylation foundation model! DNA Methylation is key for gene expression and chromatin structuring, with CpG sites often varying in diseases like cancer and aging. While important, most genomic foundation models do not capture well epigenetic and methylation signature. Introducing MethylGPT, the latest foundation model specific for DNA methylation (code available upon publishing in peer-reviewed paper)!
📌 Key Insights:
Trained on 226,555 samples (154,063 after QC & deduplication) including 49,156 curated CpG sites and 7.6B training tokens
Spans 20+ tissue types from 5,281 datasets
Disease risk prediction across 60 conditions
Robust methylation prediction (Pearson R=0.929) while maitaining performance with up to 70% missing data
Operate a Cell-Free Bio-Foundry using Large Language Models🔬
Cell-free protein synthesis systems (CFPSs) are well established platforms to synthesise a wide variety of compounds and biological molecules at scale. One roadblock to develop CFPS is to find the optimal mix of different components from cell extract. This process needs to have a deep knowledge of both cell-free and components involved and can be time consuming and expensive. Optimised production yield needs to be carried on for each specific system and for each protein of interest produced using these systems, requiring months of manual work and coding to set up automated systems.
In this paper the authors explores the untapped potential of LLMs to automate Design-Build-Test-Learn (DBTL) cycle and optimise cell-free biofoundries. All code and scripts available in the paper and are open-source!
📌 Key Insights:
Use of multiple “modules”:
SAMPLER: Generates an initial sampling of volume combinations to
be tested
DESIGNER: Converts the cell-free samples into an intermediate representation of source and destination microplates
INSTRUCTOR: Translates the destination plate data into ECHO instructions.
EXPERIMENTER: Loads the instructions from previous modules into an automated lab (ECHO 650 Liquid Handler robot)
LEARNER: Uses the protein production of each well (sample) to learn their components' interaction and suggests new experiments to run.
Reduce time to code and create the system from 2 months to 1 week
Achieved a 9-fold increase of production
QM9star: Large dataset to advance quantum chemical insights 💿
Chemical reactivity involves complex molecular transformations, often involving intermediates like ions and radicals. While experimentally challenging to characterize, understanding these species is crucial for predicting reaction outcomes. Introducing QM9star, a comprehensive dataset derived from QM9, featuring more than 1.8 millions of ions/cations/radicals with detailed chemical-physical properties to aid in machine learning studies related to chemical intermediates.
📌 Key Insights:
A rich database containing 120,280 neutral molecules, 435,669 cations, 721,441 anions, and 731,416 radicals, all at their equilibrium state and comprising the following atom types: C/H/N/O/F.
Provides global (energies, orbitals, and frequencies, polarizability) and local (coordinates and forces, formal charge and spin, and charge and spin densities) features.
Can be used in future work with machine learning models to better characterize the properties of unstable molecules in molecular reactivity.
Did you find this newsletter insightful? Share it with a colleague!
Subscribe Now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website