AI in Life Science: Weekly Insights
Weekly Insights | February 18, 2025
In this issue:
Welcome back to your weekly dose of AI news for Life Science!
This week, we have some exciting new models lined up for you:
Dive into these game-changing innovations and explore how they are transforming the biotech and healthcare landscapes!
IntegrAO: Integrate Any Omics 🧬
Another excellent framework from Bo Wang’s lab! IntegrAO enables integration of multiple omics data types, using a network approach to encode patient data into embeddings and an unified graph. These embeddings can then be used for other tasks, such as prediction of patients subtypes. The authors trained the model on a large dataset of oncology data, then demonstrated its superior performance when predicting patient subtypes in AML and other cancer types. The framework can be adapted to other data types, and fine tuned to other tasks, such as to cell subtyping, drug discovery, biomarker identification, precision nutrition, and more.
🔨 Applications:
Prediction of patient subtypes from multi-omics data. However, the framework could be applied and adapted to other prediction tasks, such as cell subtyping, drug discovery, biomarker identification, precision nutrition, and more.
📌 Key Insights:
Enables integration of multiple omics data types, such as RNASeq expression, Copy Number Variations, DNA Methylation, and more. In the article, the authors demonstrated its use for prediction of patient subtypes in cancer datasets, but the framework can be used for other data types and tasks.
Demonstrates robust performance on heterogeneous, incomplete datasets from multiple cancer cohorts. Maintains accuracy despite varying degrees of missing omics
The authors used IntegrAO to predict patient subtypes for multi-omics datasets. The method outperforms state-of-the-art tools (NEMO, MSNE) across multiple evaluation scenarios.
MEDTOK: A Multimodal Tokenizer for AI-Powered Medical Code Processing 💻
Medical AI models rely on structured electronic health records (EHRs), but traditional tokenization methods treat medical codes as isolated text, missing crucial hierarchical and relational information. Introducing MEDTOK, a multimodal medical code tokenizer, changes this by integrating both text descriptions and graph-based relationships from biomedical ontologies. By doing so, it improves EHR model performance across clinical and operational tasks, enhancing diagnosis classification, risk prediction, and drug recommendation
🔨 Applications:
Optimising AI models in healthcare by improving tokenization of medical records, leading to better predictive accuracy in clinical tasks.
📌 Key Insights:
Multimodal Encoding: Combines text embeddings from medical code descriptions with graph representations of relationships like disease co-occurrences and drug interactions.
Higher Predictive Performance: Improves AUPRC scores across all EHR models, including +11.3% on drug recommendations, a critical AI application in clinical decision-making.
Better Handling of Medical Ontologies: Captures hierarchical relationships across multiple coding systems (ICD, SNOMED, RxNorm, ATC), improving cross-system interoperability.
Seamless Integration: Can be used with any transformer-based AI model, including those for disease progression modelling, medical question-answering, and risk stratification.
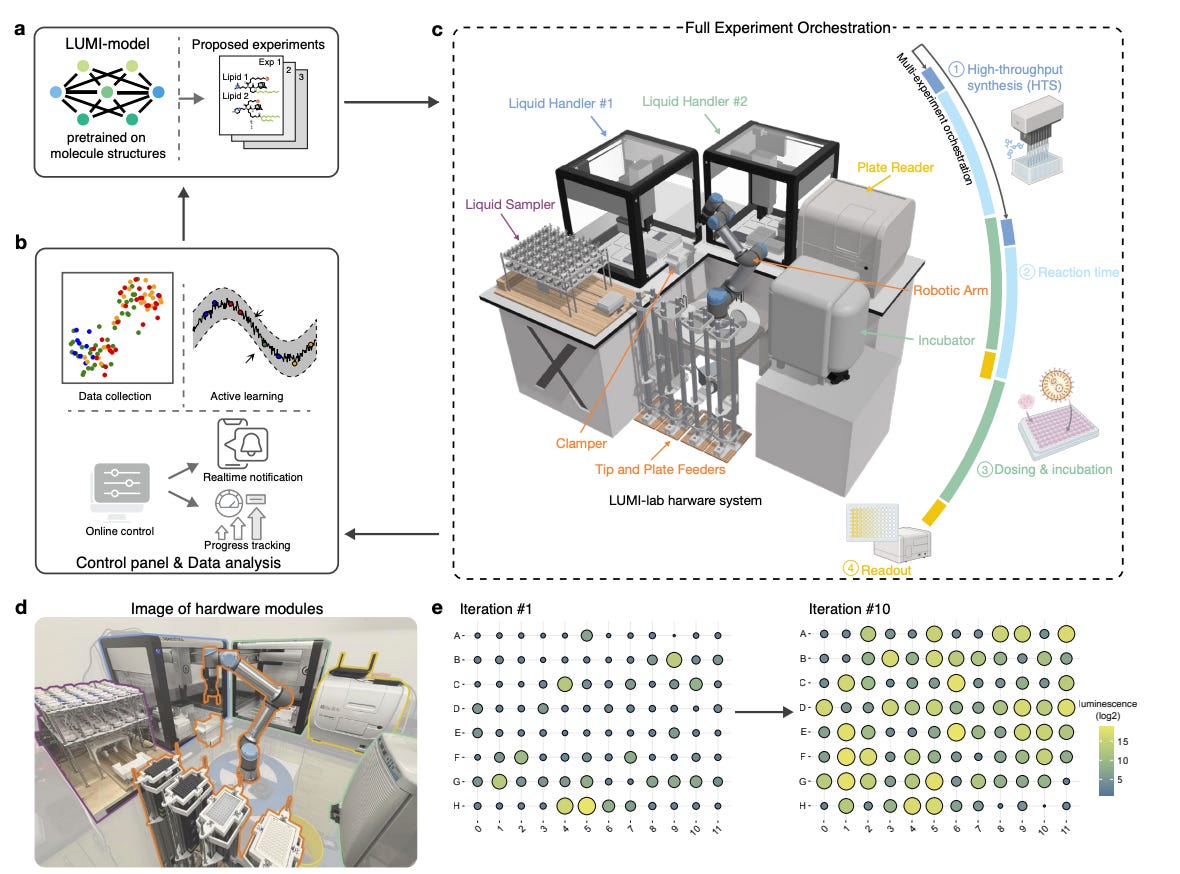
LUMI-Lab: Self driving lab for mRNA delivery🔬
The complexity of molecular discovery requires autonomous systems that efficiently explore vast and uncharted chemical spaces. While integrating artificial intelligence (AI) with robotic automation has accelerated discovery, its application remains constrained in fields with scarce historical data. One such challenge is the design of lipid nanoparticles (LNPs) for mRNA delivery, which has relied on expert-driven design and is hindered by limited datasets. Introducing LUMI-lab, a self-driving lab (SDL) system that enables efficient learning with minimal wet-lab data by integrating a molecular foundation model with an automated active-learning experimental workflow
🔨 Applications:
Streamline development of LNPs for mRNA drug delivery
📌 Key Insights:
LUMI-lab synthesised and evaluated over 1,700 LNPs, identifying ionisable lipids with superior mRNA transfection potency in human bronchial cells compared to clinically approved benchmarks.
The top-performing lipid, LUMI-6, achieved 20.3% gene editing efficacy in lung epithelial cells in murine models, surpassing the highest efficiency reported for inhaled LNP-mediated CRISPR-Cas9 delivery in mice to date.
Brominated lipids autonomously identified as a novel structural feature that enhances mRNA transfection—an insight previously unrecognised in LNP design
Did you find this newsletter insightful? Share it with a colleague!
Subscribe Now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website