AI in Life Science: Weekly Insights
Weekly Insights | Jun 10, 2025
In this issue:
Welcome back to your weekly dose of AI news for Life Science!
This week, we have some exciting new models lined up for you:
Boltz-2: Supercharging Drug Discovery with AI-Powered Affinity Predictions💊
KRONOS : Unlocking Cellular Secrets with Spatial Proteomics 🔬
Dive into these game-changing innovations and explore how they are transforming the biotech and healthcare landscapes!
Boltz-2: Supercharging Drug Discovery with AI-Powered Affinity Predictions 💊
Accurately predicting how tightly a drug-like molecule will bind to its protein target is a central challenge in biology and medicine. While recent AI models have gotten better at predicting the structure of these interactions, they have struggled to predict binding affinity—a critical factor for a drug's effectiveness. Researchers have now developed Boltz-2, a new AI foundation model that excels at predicting both molecular structure and binding affinity. The key result is that Boltz-2 is the first AI model to approach the accuracy of gold-standard physics-based methods called free-energy perturbations (FEP), while being over 1,000 times more computationally efficient.
🔨 Applications:
Accelerate hit-to-lead optimization by ranking analogue series with FEP-like accuracy (R = 0.66) in ~20 GPU s instead of multi-hour simulations.
Enable large-scale virtual screening, boosting enrichment 18.4-fold in the top 0.5 % of compounds on the MF-PCBA benchmark.
Discover novel binders: paired with the SynFlowNet generator, Boltz-2 produced ten synthesizable TYK2 inhibitors, all predicted as tight binders by ABFE.
Improve dynamic structural studies by lifting RMSF Pearson correlations up to 0.82 on ATLAS molecular-dynamics datasets, outperforming specialized models.
📌 Key Insights:
Lightning-fast, lab-level accuracy – Boltz-2 delivers predictions in seconds yet comes within reach of gold-standard FEP accuracy, running >1,000 × quicker than those multi-hour physics simulations.
Sharper screening – When sifting millions of molecules, Boltz-2 pulls about 18 × more true hits into the very top slice of ranked candidates than traditional docking, saving time and lab costs.
Sees proteins breathe – Its predictions mirror how proteins flex in molecular-dynamics data, giving scientists a clearer picture of tricky, flexible drug targets.
Plays nicely with AI design tools – Hooked up to a molecule generator, Boltz-2 helped craft new TYK2 inhibitor ideas that later high-precision simulations confirmed as strong binders, pointing to real drug leads.
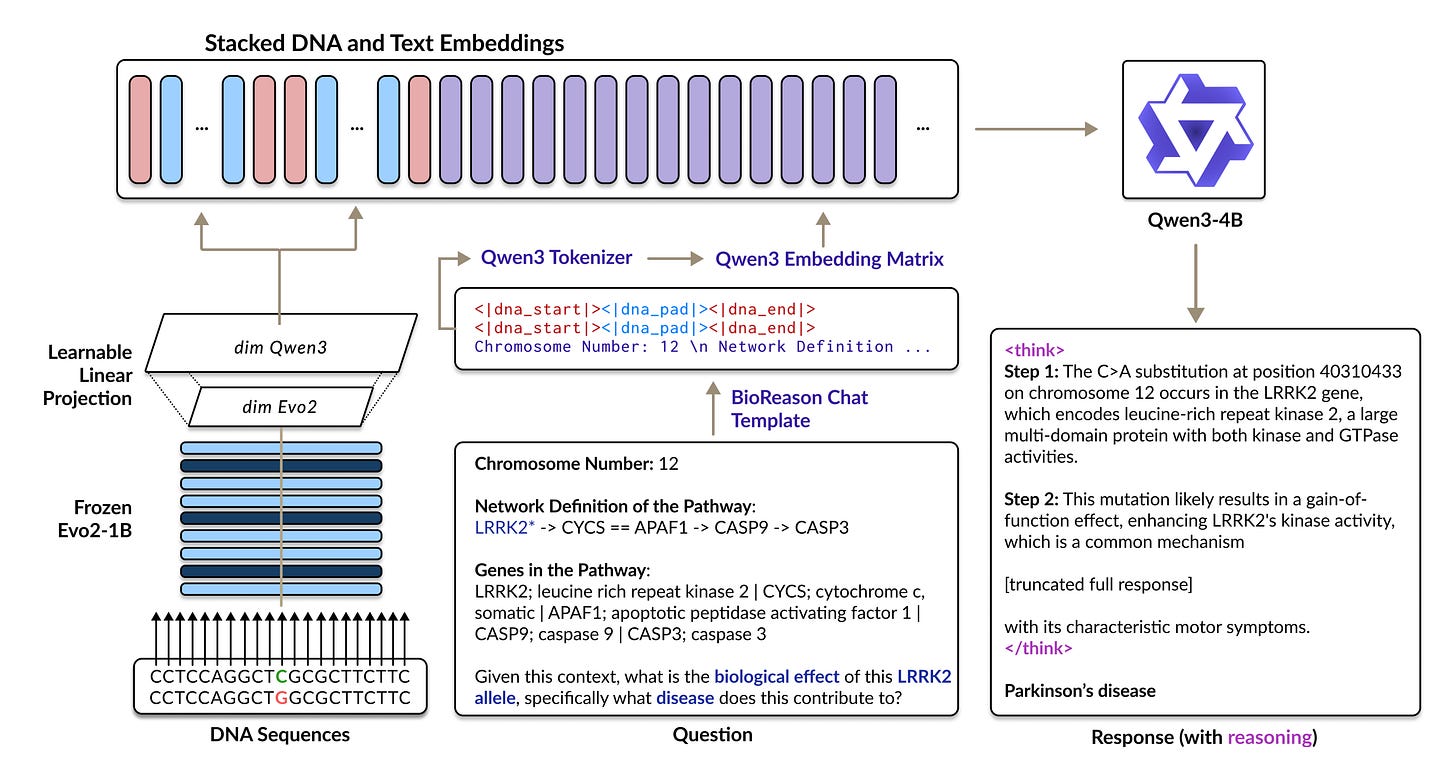
BIOREASON: Inverse‑Folding Nanobody Sequence Design 🧬
Unlocking deep, understandable insights from complex genomic data is a significant challenge in scientific discovery. Current DNA foundation models, while powerful, often function as "black boxes" that struggle with multi-step reasoning , and large language models (LLMs) cannot effectively process raw genetic sequences on their own. Researchers have introduced BIOREASON, a pioneering architecture that solves this problem by deeply integrating a DNA foundation model with an LLM, enabling the system to reason directly with genomic information. This novel approach led to an average 15% performance gain over strong single-modality baselines and improved disease pathway prediction accuracy from 88% to 97%.
🔨 Applications:
Accelerate biological discovery by offering deeper, mechanistic insights into how genomic data translates to biological function.
Enable researchers to form new, testable scientific hypotheses by providing transparent, step-by-step explanations for its predictions.
Improve the understanding of complex disease pathways and variant effects, with future applications in clinical mutation interpretation and genome-wide association studies (GWAS).
Discover nuanced biological patterns by leveraging a system that synergistically combines the sequence representation power of DNA models with the sophisticated reasoning of LLMs.
📌 Key Insights:
Unified framework: Novel multimodal architecture that lets LLMs directly process genomic sequences as input
Strong disease prediction: BIOREASON achieves higher accuracy compared to LLM or DNA language models alone in predicting disease from gene pathways. Consistent 15%+ performance gains over DNA-only or LLM-only baselines
Strong interpretability: Combination of LLM and DNA language model provide mechanistic biological insights, bridging the gap between "black box" DNA foundation models and interpretable scientific reasoning.
KRONOS : Unlocking Cellular Secrets with Spatial Proteomics 🔬
Spatial proteomics, a set of technologies that map protein locations at single-cell resolution, has been held back by analysis methods that struggle to scale and often miss the bigger picture. To solve this, researchers have developed KRONOS, a new foundation model built specifically for spatial proteomics. KRONOS is a Vision Transformer trained on a massive and diverse dataset of over 47 million multiplexed image patches. The model demonstrates state-of-the-art performance across a variety of critical tasks, including cell classification and patient stratification, proving its ability to learn biologically rich representations from complex tissue images.
🔨 Applications:
Accelerate cell phenotyping with high accuracy, even with limited labeled data.
Enable new segmentation-free analysis workflows by processing image patches directly, bypassing potential errors from cell segmentation.
Improve patient stratification by predicting clinical outcomes, such as treatment response, from tissue images.
Discover similar biological structures across different studies and datasets using a powerful reverse image search engine for spatial patterns.
📌 Key Insights:
Superior Performance: In cell phenotyping tasks, KRONOS consistently outperformed other models. For instance, on the classical Hodgkin's Lymphoma (cHL) dataset, it achieved a balanced accuracy of 0.7358±0.0089, significantly surpassing models like DINO-v2 (0.6210±0.0121) and UNI (0.5570±0.0136).
Exceptional Data Efficiency: KRONOS is highly effective even with minimal supervision. In a few-shot learning test, KRONOS trained with only 100 labeled cells per class achieved a balanced accuracy (0.7143±0.0410 on the DLBCL-1 dataset) that significantly outperformed other models trained with ten times more data.
Robust Generalization: The model's representations are highly transferable across different datasets and conditions. When trained on one lymphoma dataset (DLBCL-1) and tested on another (DLBCL-2), KRONOS achieved a balanced accuracy of 0.7896±0.0072, demonstrating its ability to handle batch effects.
Novel Architecture: KRONOS introduces key adaptations for multiplexed imaging, such as a dedicated marker encoding system. This architectural choice was critical, yielding up to a 37.4% absolute increase in balanced accuracy on the cHL dataset compared to models without it.
Did you find this newsletter insightful? Share it with a colleague!
Subscribe Now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website