AI in Life Science: Weekly Insights
Weekly Insights | July 8, 2025
In this issue:
Welcome back to your weekly dose of AI news for Life Science!
This week, we have some exciting new models lined up for you:
ProteinMPNN + AbLang: A Simple Boost for Antibody Design Accuracy 🧬
BMFM-RNA: A Unified Platform for Building Transcriptomic Foundation Models 📊
Dive into these game-changing innovations and explore how they are transforming the biotech and healthcare space!
What’s slowing you down? Let us tackle it.
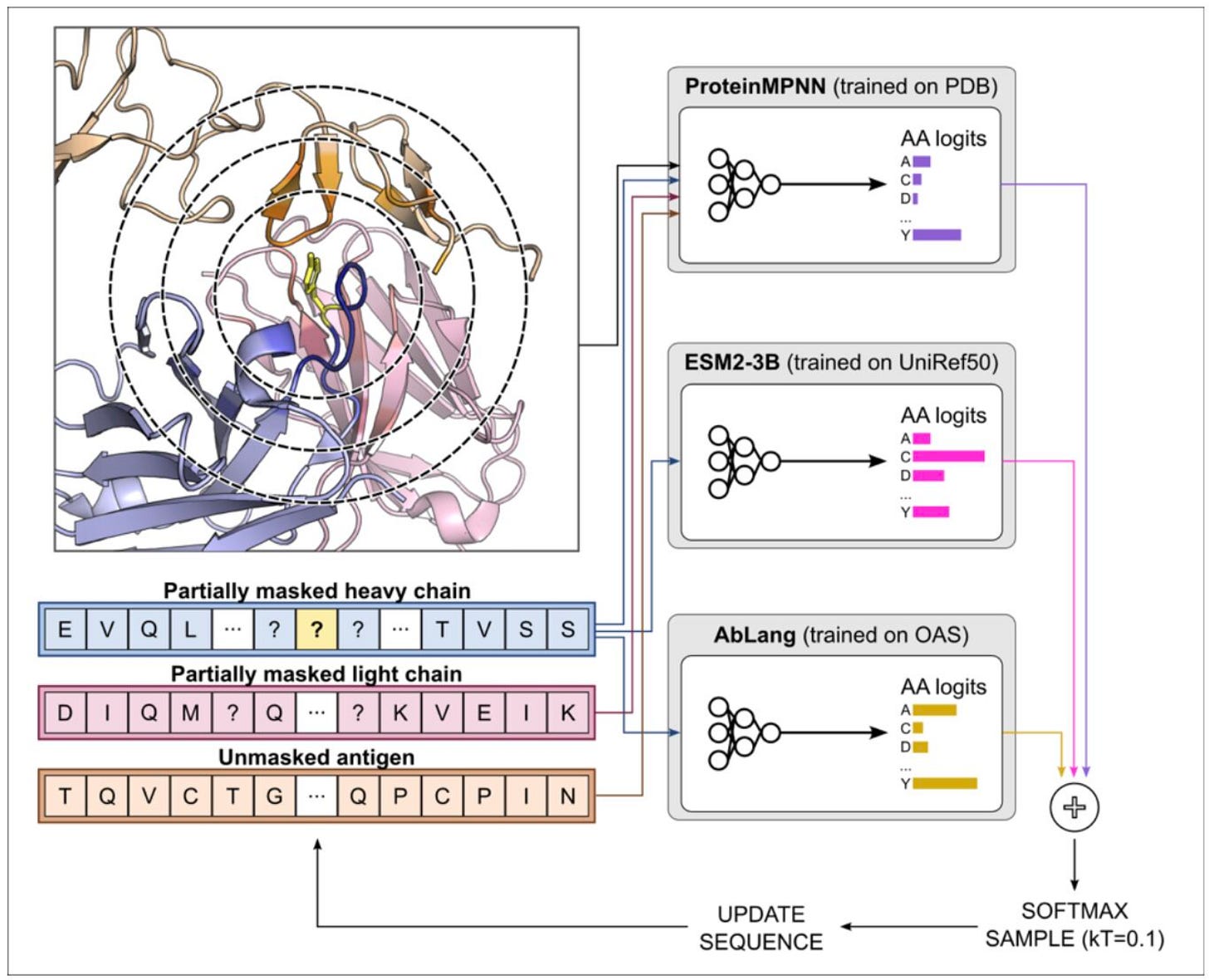
ProteinMPNN + AbLang: Boosting Antibody Design Without Retraining 🧬
Designing functional antibodies requires precise control over complementarity-determining regions (CDRs), the loops that bind antigens. While structure-based tools like ProteinMPNN excel at designing well-folded proteins, they often struggle to generate realistic antibody CDRs, especially when working with limited screening capacity. To tackle this, researchers at GSK combined ProteinMPNN with AbLang, a language model trained on millions of antibody sequences, to guide designs toward native-like, functional antibodies, all without retraining or building new models.
By adding AbLang’s sequence bias to ProteinMPNN’s structure predictions, the combined method generates designs that are more consistent with natural antibodies and show significantly higher experimental success rates.
🔬 Applications
Higher-Quality Antibodies - Produces CDR loops that better match germline V-genes, improving the developability of designed antibodies for therapeutic use.
Improved Success with Fewer Variants - Enables practical lab-scale workflows by raising the hit rate for functional binders, reducing the need for costly large-scale screens.
Seamless Integration - Works as a plug-in layer for existing ProteinMPNN pipelines, no retraining required, just a simple ensemble during inference.
📌 Key Insights
Antibody-Like Sequences - The combined method raises V-gene sequence identity from 75% with ProteinMPNN alone to levels matching the ~90% median seen in approved therapeutic antibodies.
10× Higher Hit Rate - In head-to-head lab tests redesigning trastuzumab’s CDRH3, only 3 of 96 ProteinMPNN designs bound HER2, compared to 36 of 96 with the ProteinMPNN + AbLang ensemble.
Simple Ensemble, Big Impact - Delivers the benefits of fine-tuned antibody design without retraining, showing clear improvements in sequence recovery and structural refolding accuracy.
BMFM-RNA: A Unified Platform for Building Transcriptomic Foundation Models 📊
Single-cell transcriptomics has transformed how we study gene expression and cell identity, but foundation models for scRNA-seq often differ wildly in how they’re built, making it hard to benchmark methods or combine the best ideas. BMFM-RNA is a new open framework that standardizes how researchers build, train, and compare transcriptomic foundation models. Its modular design includes a new pretraining strategy, the Whole Cell Expression Decoder (WCED), which improves on prior methods by better capturing global expression patterns. With BMFM-RNA, researchers can systematically test combinations of masking, multi-task learning, and new objectives therefore driving more robust, interpretable models for single-cell biology.
🔨Applications
Flexible Design - Build and benchmark models for tasks like cell type annotation, batch correction, perturbation prediction, and gene regulatory network inference using a single configurable pipeline.
Better Data Efficiency - Achieve competitive or superior clustering and annotation results with only 1–10% of CELLxGENE data, reducing compute while maintaining state-of-the-art performance.
Community Foundation - BMFM-RNA is fully open-source, enabling teams to reproduce published baselines, extend objectives, and accelerate robust transcriptomic model development.
📌 Key Insights
WCED Outperforms Standard Objectives - The new Whole Cell Expression Decoder (WCED) objective delivered an average zero-shot clustering score of 0.756, outperforming scGPT (0.719) and other baselines on 12 benchmark datasets.
Less Data, Stronger Results - Using just 10% of CELLxGENE, WCED-based models achieved up to 3.7 percentage points higher accuracy for cell type clustering and annotation than models trained on full datasets, highlighting BMFM-RNA’s data efficiency.
Robust Annotation on Real Data - In fine-tuning tests, WCED models reached up to 89% accuracy on challenging disease datasets like Multiple Sclerosis, beating scGPT’s 85%, and delivered higher F1 scores for rare cell type detection.
g-xTB: DFT Accuracy at Tight-Binding Speed ⚡
Density functional theory (DFT) is the standard tool for accurate quantum chemical simulations, but it’s too computationally demanding for very large systems or high-throughput workflows. Semiempirical tight-binding methods like GFN2-xTB help bridge this gap but still have well-known accuracy limits. The new g-xTB method, developed by the Grimme group, pushes tight-binding accuracy much closer to high-level DFT while keeping the speed advantage. Covering all elements up to actinides, g-xTB is designed to be a drop-in replacement for its predecessors and a robust alternative to mid-level DFT for many real-world applications.
🔨Applications
Large-Scale Screening - Enables fast yet accurate calculations for thermochemistry, conformer searches, non-covalent interactions, and reaction barriers, critical for high-throughput studies and big molecular datasets.
Better for Metals and Complex Systems - Improves reliability for transition-metal complexes, charged species, and spin states, areas where previous SQM and TB methods struggled.
Robust Drop-In Tool - Minimal empirical tuning and a wide chemical training set make g-xTB highly transferable, a general-purpose tool ready for practical workflows across chemistry, materials, and catalysis.
📌Key Insights
Half the Error, Same Speed - On the GMTKN55 benchmark, g-xTB cuts the mean absolute deviation to 9.3 kcal/mol, half that of GFN2-xTB (25 kcal/mol) and matching the performance of efficient composite DFT (WTMAD-2 ≈ 9).
Proven on 32,000 Cases - Extensively benchmarked on ~32,000 energies, g-xTB shows big improvements for thermochemistry, conformer energies, and transition-metal barriers, outperforming GFN2-xTB by 2–3× on reaction barrier sets.
Minimal Overhead - Delivers these gains with only 30–50% more compute time than GFN2-xTB, still ~750× faster than low-cost DFT, making it practical for large-scale jobs.
Did you find this newsletter insightful? Share it with a colleague!
Subscribe Now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website