AI in Life Science: Weekly Insights
Weekly Insights | March 2, 2025
In this issue:
Welcome back to your weekly dose of AI news for Life Science!
This week, we have some exciting new models lined up for you:
Dive into these game-changing innovations and explore how they are transforming the biotech and healthcare landscapes!
If you are interested to learn how you can connect all these tools, reach out to us!
Evo-2: Genome modelling at scale 🧬
The Arc institute presents Evo-2, a massive DNA foundation model trained on more than 9.7 T nucleotides from 100,000 genomes from different species. It represents a leap forward compared to the previous model, called Evo, published just one year ago. Despite being trained only on DNA sequences, without providing any genomic annotation, Evo 2 learned gene regulatory patterns and mutation effects by itself.
🔨 Applications:
Design of synthetic genomes - Design new (small) synthetic genomes for specific use cases
Annotation of genomic regions - Identify regulatory elements, exon-intro structures, and understand the gene and lncRNA essentiality
Clinical Variant interpretation - Classify variant of uncertain significant into pathogenic e.g. for BRCA1 and BRCA2 mutations
📌 Key Insights:
Evo 2 is trained on OpenGenome2, a collection of DNA sequences from more than 100,000 species, including Bacteria, Archaea, Eukaryotes, and bacteriophages. The input does not include any annotation, and the model learns genomic structures, protein coding effects and mutational effects by itself.
Evo2 has a context size of 1 million nucleotides, meaning that it is able to model interactions between DNA sequences that are far from each other in the genome. The previous Evo model had a context of just 131K, and other models based on the transformer architecture struggle to reach 100K.
Training a model of this size on DNA sequences is a technical challenge. One key feature to achieve such task is the adoption of the Hyena 2 architecture, which has several advantages compared to the classic transformer. The authors released a companion paper to describe how to optimise this task.
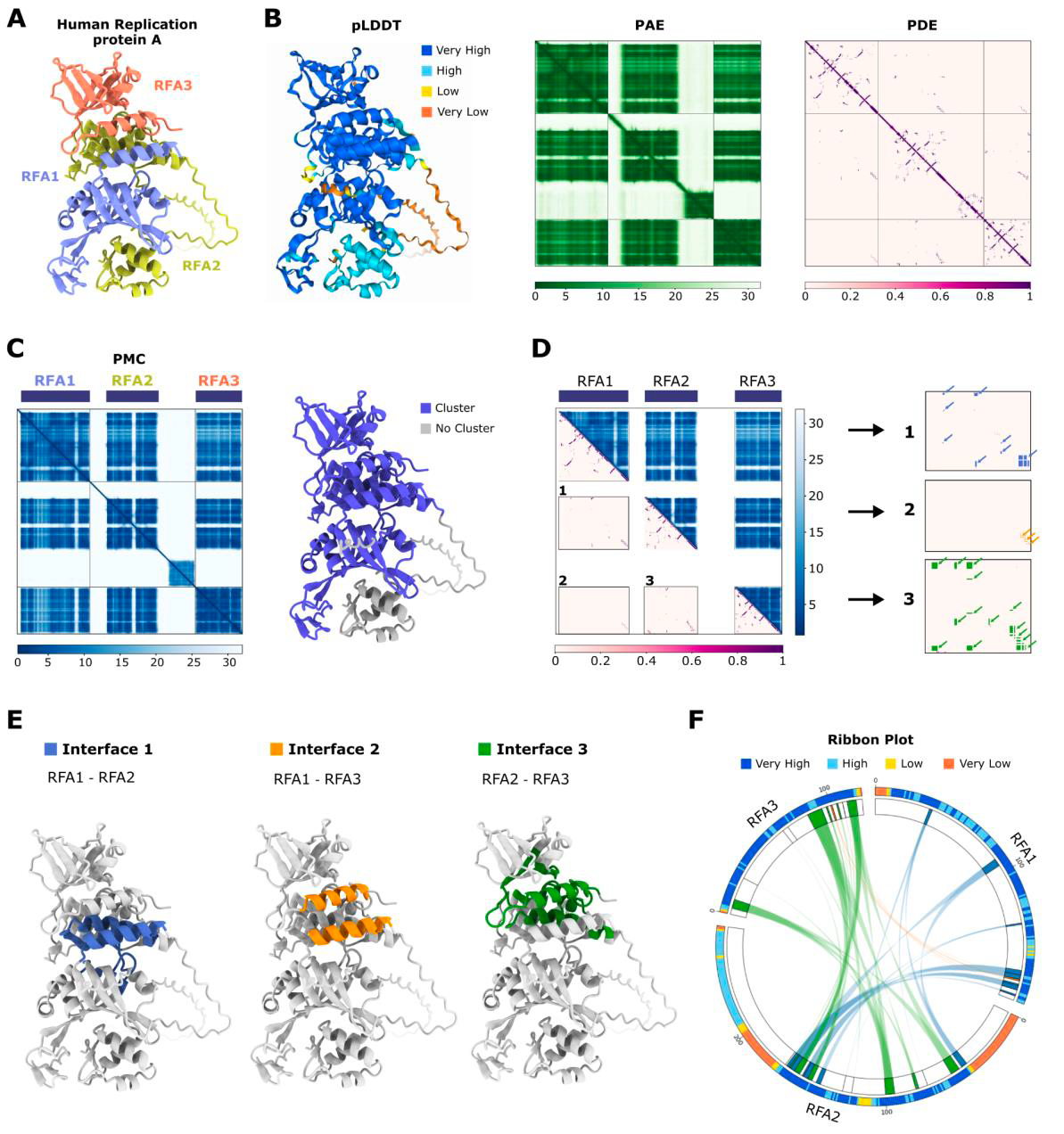
AlphaBridge: Visualising macromolecule interactions 💻
The complexity of macromolecular structure prediction requires innovative approaches to efficiently explore vast interaction spaces. While AI-powered methods like AlphaFold have revolutionised protein structure prediction, their application to macromolecular complexes presents new challenges and opportunities. This study introduces AlphaBridge, a novel tool that leverages AlphaFold3's prediction matrices in a graph-based community clustering approach to analyze and visualize macromolecular interactions. By processing these "interaction images" through multidimensional algorithms, AlphaBridge generates chord diagrams that summarize predicted interfaces, including confidence scores and sequence conservation. This method offers an efficient framework for screening and ranking predicted interactions, potentially accelerating the discovery and analysis of complex macromolecular structures in life sciences
🔨 Applications:
Drug Discovery & Therapeutic Targeting – Identifies key protein interactions to aid in drug development and precision medicine.
Genetic & Evolutionary Studies – Provides insights into evolutionary conservation and the effects of mutations on protein interactions.
📌 Key Insights:
Offer an efficient framework for screening and ranking predicted interactions, prior to more detailed analysis
Easy-to-use script find_interface on GitHub, with conda environment and only one line to enter the terminal
Demonstrations with data representation on large complex biological systems
ProtGPS: Understanding protein location form amino acid sequences 🌍
Proteins function within specific regions of a cell, and their correct localization is essential for proper cellular activity. Previously, scientists believed that proteins reached these compartments primarily through structural mechanisms. However, recent research has revealed that a protein’s precise localisation is just as crucial as its function, as mislocalisation is linked to severe diseases, including neurodegenerative disorders and various cancers. Introducing ProtGPS—a deep-learning model that accurately predicts the subcellular destinations of human proteins, offering new insights into cellular organisation and disease mechanism.
🔨 Applications:
Disease Research & Therapeutics – Identifies mutations that mislocalise proteins, aiding in understanding diseases (e.g., cancer, neurodegeneration) and developing targeted drug therapies.
Synthetic Biology & Protein Engineering – Enables design of custom proteins with precise subcellular localisation for improved metabolic pathways, gene therapy, and industrial biotech applications.
📌 Key Insights:
Trained on more than 5,500 human proteins. It takes amino acid sequences as input and makes predictions about the subcellular localisation of a protein by identifying several patterns that determine its suitability to various compartments.
Reached accuracy scores from 83% to 95% across the partitions of a cell in 12 compartments.
Trained on 200k+ proteins with disease-associated mutations, predicting where those mutated proteins would localize. This demonstrates the value of ProtGPS as a tool for generating functional proteins, understanding disease, and identifying new therapeutic avenues.
Did you find this newsletter insightful? Share it with a colleague!
Subscribe Now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website