AI in Life Science: Weekly Insights
Weekly Insights | August 5, 2025
In this issue:
Welcome back to your weekly round-up of new tools and methods in life sciences research.
This week, we’re spotlighting three innovations that advance retrosynthesis, antimicrobial prediction, and 3D pathology triage:
RSGPT: Large-Scale Language Model for Retrosynthesis Planning🧪
ApexOracle: Foundation Model for Antimicrobial Prediction and Molecule Design 💊
Each of these tools addresses long-standing bottlenecks in biomedical workflows, from accelerating synthesis planning and guiding antibiotic design to improving clinical decision-making in volumetric histopathology.
What’s your most time consuming task along the drug discovery process?
We will send you open-source tools specific to your pain point.
RSGPT - Large-Scale Language Model for Retrosynthesis Planning 🧪
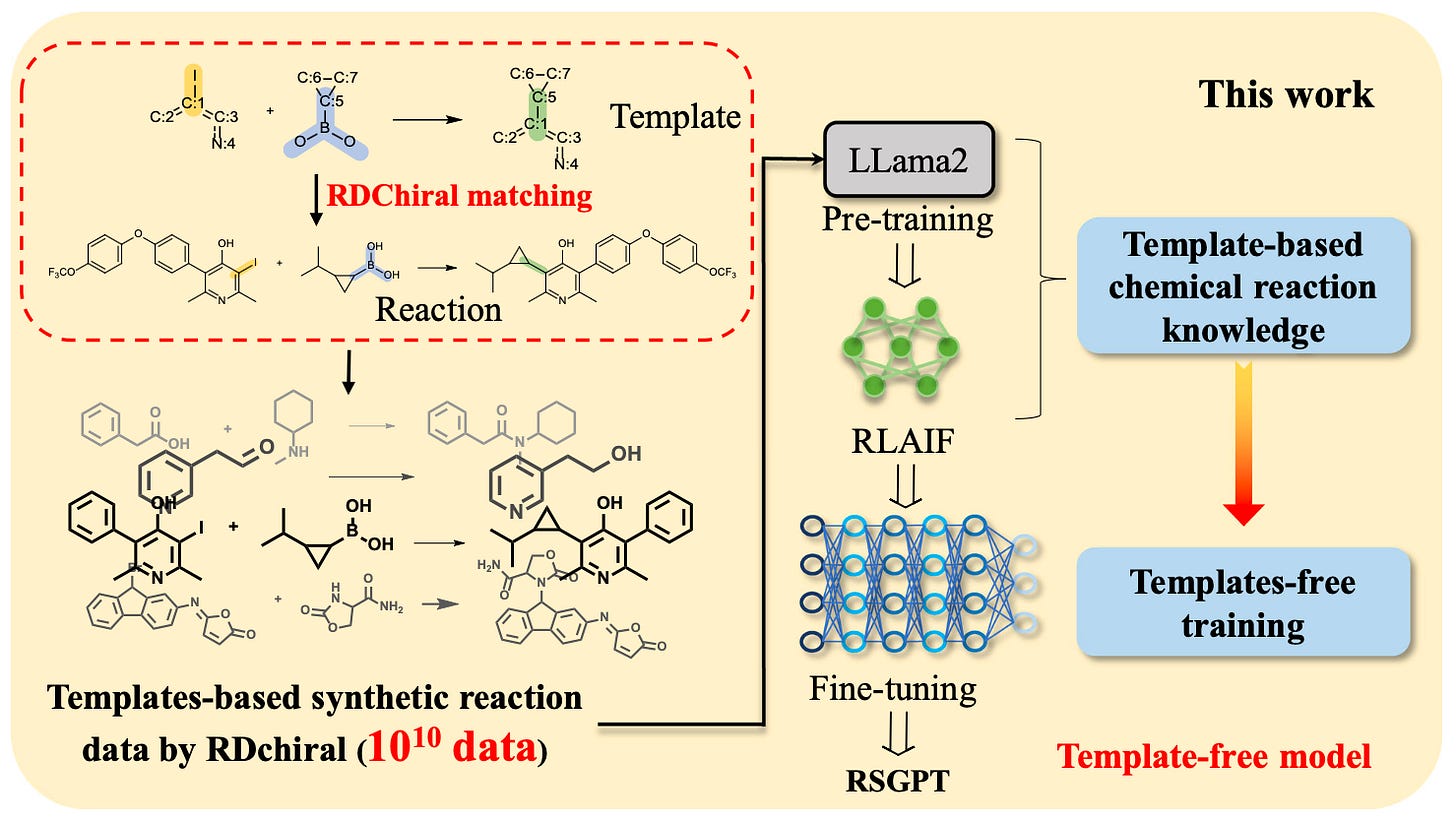
Retrosynthesis prediction traditionally requires expensive template engineering or large reaction databases with atom mapping. RSGPT introduces a transformer-based, template-free framework trained on over 10 billion synthetically generated reactions using SMILES strings. It incorporates reinforcement learning from AI feedback (RLAIF) and aggressive data augmentation to learn rich chemical priors directly from data.
The model achieves 77.0 percent Top-1 accuracy on USPTO-50k with augmentation, 63.9 percent on USPTO-MIT, and 59.2 percent on USPTO-FULL, surpassing existing state-of-the-art template-free and semi-template models. Its high SMILES validity rate (97.7 percent) and Tanimoto similarity (0.840) further underscore its reliability.
🔬 Applications
Template-Free Planning - Enables fast and flexible retrosynthesis predictions without the need for expert-crafted reaction rules.
Scalable Drug Discovery - Supports multi-step synthesis of clinical molecules including Osimertinib and Vonoprazan.
Diverse Reaction Coverage - Trained on over 10 reaction classes and generalises well across noisy and clean datasets.
📌 Key Insights
SOTA Top-1 Accuracy - Outperforms prior methods with Top-1 accuracy of 77.0 percent on USPTO-50k with 20x augmentation.
High Validity and Similarity - Achieves 97.7 percent SMILES validity and 0.840 average Tanimoto similarity to ground truth outputs.
Generalisation Across Benchmarks - Retains high performance across
USPTO-MIT and FULL datasets, improving over R-SMILES by up to 4.7 percentage points.
ApexOracle - Foundation Model for Antimicrobial Prediction and Molecule Design 💊
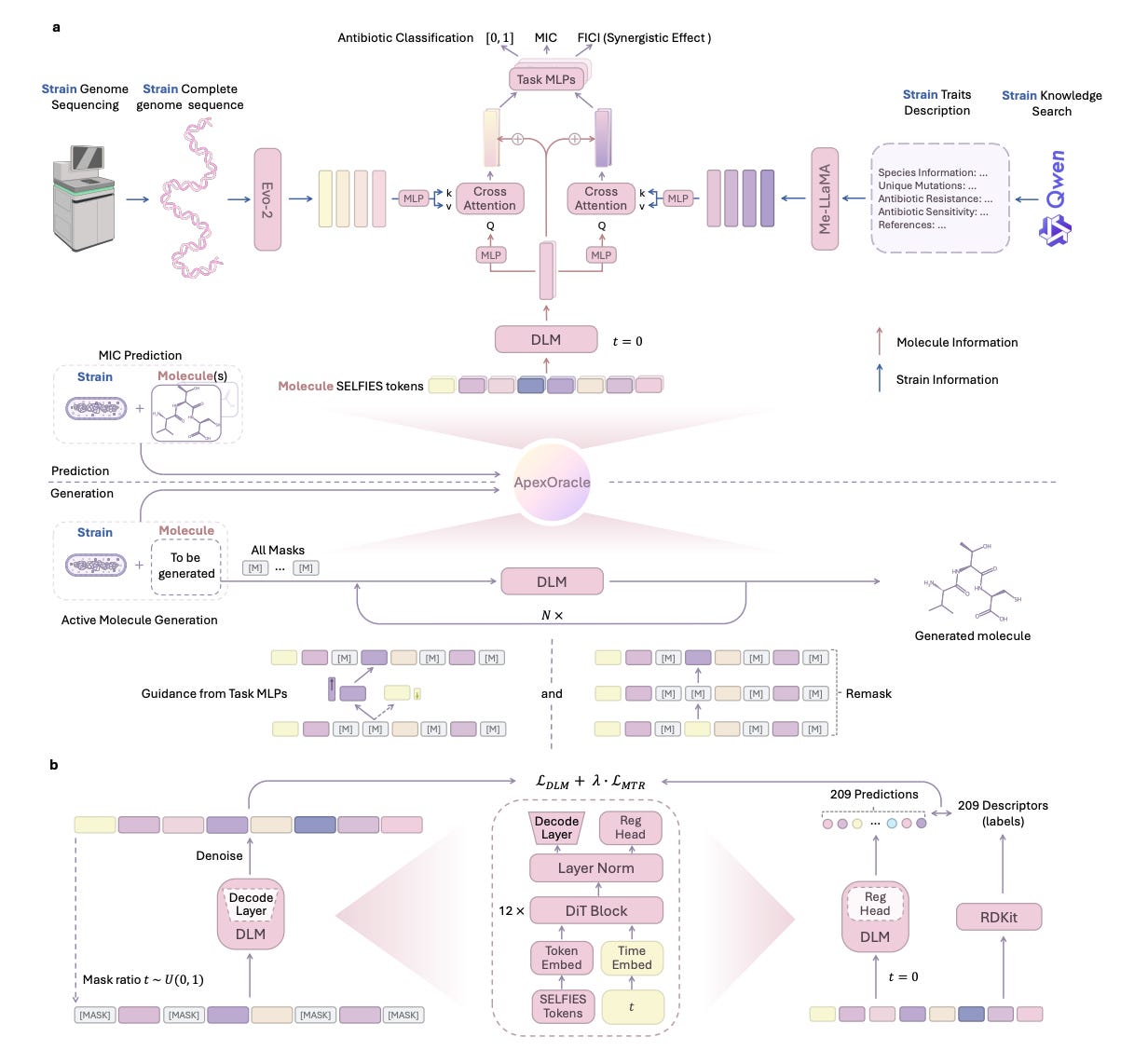
Predicting antibiotic activity and synergy across pathogen strains is limited by the diversity and sparsity of existing datasets. ApexOracle introduces a multi-modal foundation model that integrates SELFIES-based chemical language modeling with pathogen genomics and curated phenotype data. It is trained on over 67,000 AMP-strain activity pairs and 121 million unique molecules.
In antibiotic prediction tasks, ApexOracle delivers 8.3 percent AUROC and 37.7 percent AUPRC gains over fine-tuned baselines. For antimicrobial synergy, it achieves a mean AUROC of 0.7539 across 2732 interactions, showing strong zero-shot transfer and generalisation across phylogenetically distant species.
🔬 Applications
Antimicrobial Prediction - Predicts MIC values for unseen strains without requiring strain-specific training data.
Combination Screening - Accurately models synergistic effects between AMPs and small molecules to reduce resistance emergence.
Strain-Agnostic Design - Guides de novo antimicrobial generation with generalisable strain-aware molecular embeddings.
📌 Key Insights
Zero-Shot Superiority - Outperforms fine-tuned baselines in zero-shot small molecule classification across 3 pathogen strains.
Multi-Species Performance - Maintains an average R2 of 0.4337 across 11 taxonomic clusters in species-wise evaluation.
Synergy Prediction - Accurately classifies AMP combination outcomes with AUPRC of 0.7454 using only 2732 data points.
CARP3D - Context-Aware Risk Prediction in 3D Pathology 🧠
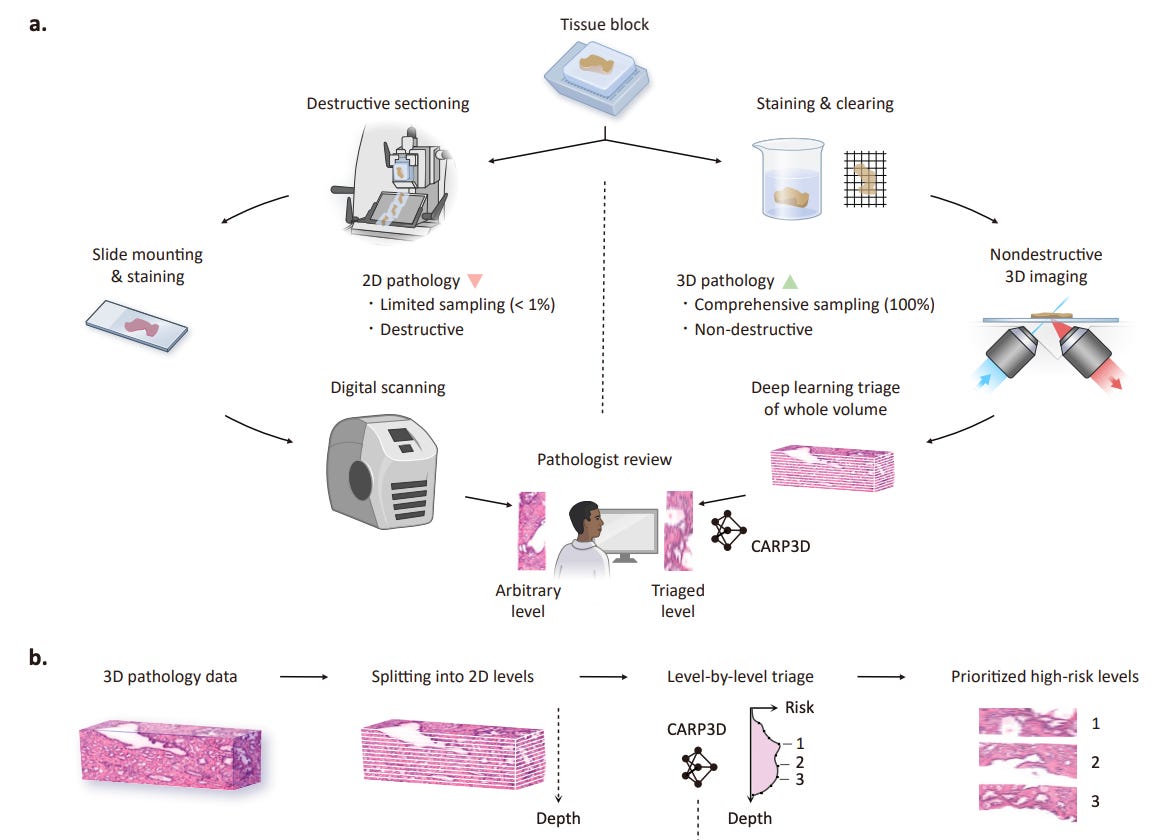
Standard pathology relies on sparse 2D sections, missing critical features in complex 3D tissue structures. CARP3D introduces a 2.5D multiple instance learning pipeline for clinical triage that identifies the highest-risk cross-sections in 3D biopsy volumes. It integrates spatial depth context using lateral and depth aggregation networks, improving the prioritisation of pathologist review.
On 3D datasets for prostate and esophageal tissue, CARP3D improved AUC from 0.871 to 0.939 for prostate and 0.895 to 0.921 for esophageal cancer detection. Balanced accuracy increased by over 7 percentage points and F2 score gains confirmed enhanced sensitivity to high-risk features.
🔬 Applications
Biopsy Triage - Flags high-risk regions in volumetric pathology data, improving diagnostic accuracy and efficiency.
Clinical Versatility - Tested on 112 prostate and 95 esophageal biopsy samples using OTLS imaging with resolution down to 0.6 µm.
Depth-Aware Prediction - Captures contextual morphology across slices, reducing spatial noise in risk prediction.
📌 Key Insights
Improved AUC - Achieves 0.939 AUC on prostate and 0.921 AUC on esophageal cases, outperforming 2D analysis baselines.
Efficient Depth Aggregation - Optimal depth range of 60 µm for prostate and 10 µm for esophagus, using three stacked slices.
Pathologist Validation - Enhances human diagnostic accuracy through AI triage without altering final decision-making workflow.
Did you find this newsletter insightful? Share it with a colleague!
Subscribe Now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website