AI in Life Science: Weekly Insights
Weekly Insights | May 14, 2025
In this issue:
Welcome back to your weekly dose of AI news for Life Science!
This week, we have some exciting new models lined up for you:
DeepSpot: Spatial-Context‑Aware Prediction of Spatial Transcriptomics from Routine Histology 🧫🔮
PoseX: AI Docking Tops Physics Rules for Real‑World Protein‑Ligand Cross‑Docking 🧪
Dive into these game-changing innovations and explore how they are transforming the biotech and healthcare landscapes!
DeepSpot: Spatial-Context‑Aware Prediction of Spatial Transcriptomics from Routine Histology 🧫🔮
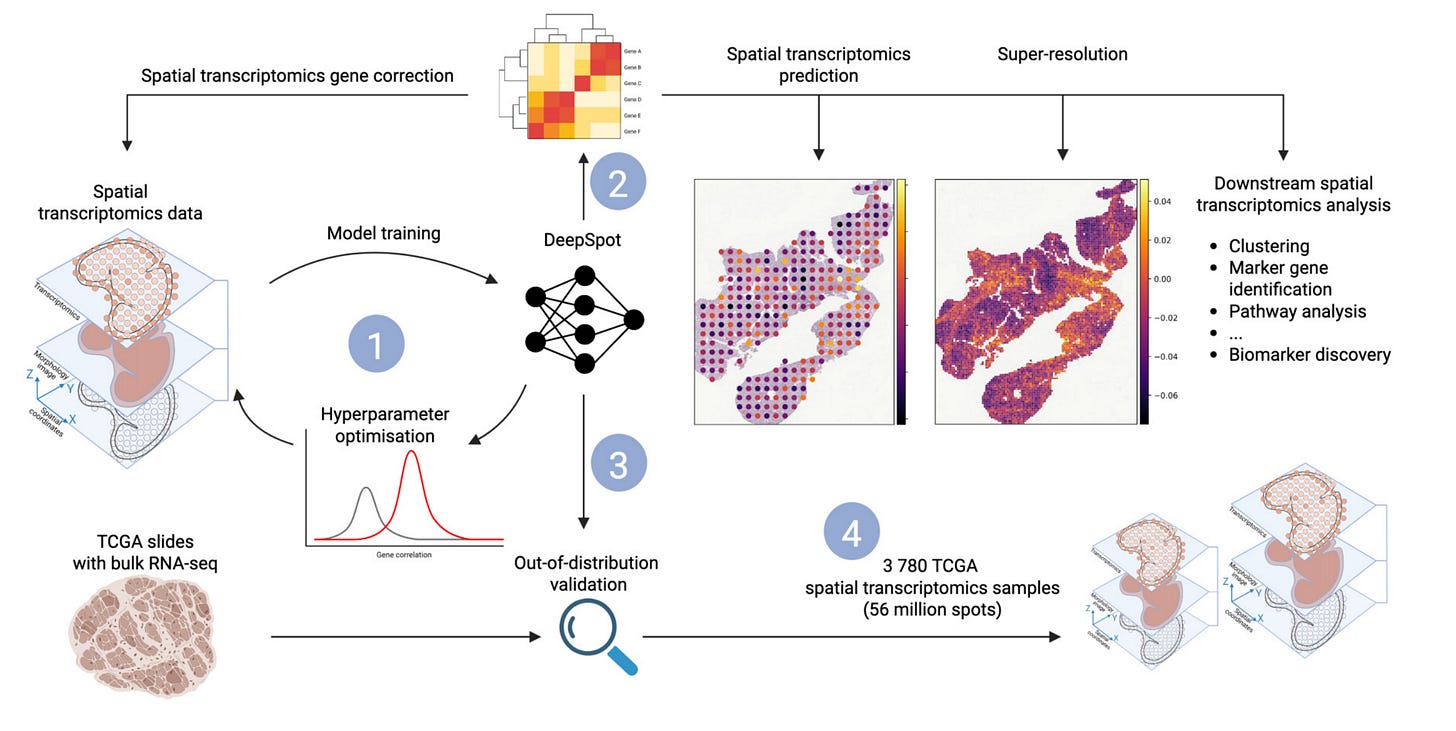
Current spatial transcriptomics (ST) platforms are costly, labor‑intensive and limited in the area they can profile, slowing the translation of spatial omics into everyday research and clinical practice. DeepSpot introduces a deep‑set neural network that fuses (1) features from large H&E foundation models (UNI, Phikon, H‑optimus‑0) with (2) fine‑grained local “sub‑spot” morphology and (3) the broader neighbourhood context of each spot. Trained to regress the 5 000 most variable genes, it markedly outperforms prior ST‑from‑H&E approaches across melanoma, kidney, lung and colon cancer cohorts.
🔨 Applications:
Augment Routine Histology – Convert standard H&E slides into spatial gene‑expression maps so any lab can glean molecular insights without extra wet‑lab steps or expensive ST kits.
Scale Up Cohort‑Level Studies – Rapidly build virtual spatial‑omics datasets for thousands of archived specimens, enabling population‑scale discovery and meta‑analysis at minimal cost.
Zoom In on Tissue Micro‑Environments – Virtually increase resolution to probe tumour–immune interfaces, germinal centres or fibrotic niches when physical high‑res ST isn’t practical.
📌 Key Insights:
Robust Performance Gains – Median Pearson r improved by 29 % on Tumor Profiler melanoma (top 500 genes) and by 77 % on single‑cell Xenium lung (top 100 genes) over the best prior methods.
Spatial Bags-of-Spots Architecture – Treats each Visium spot as a bag of nine sub‑spots plus its neighbouring spots, captured with permutation‑invariant deep sets; jointly learns local and global tissue cues.
Out‑of‑Distribution Generalisation – On 3 780 unseen TCGA slides, pseudo‑bulk profiles derived from DeepSpot ST correlate better with matched bulk RNA‑seq than previous models – and, for SKCM survival, even outperform true bulk RNA (C‑index 0.608 vs 0.556).
Rescues Low‑Quality Samples – Recovers expression of TLS marker genes in Visium slices that failed QC, demonstrating resilience to low RNA sensitivity and spot contamination.
nanoFOLD: Inverse‑Folding Nanobody Sequence Design 🦙
Traditional inverse‑folding models are trained on general proteins or full‑length antibodies, limiting their accuracy on camelid single‑domain antibodies (nanobodies). nanoFOLD fine‑tunes the ESM‑IF architecture on nanobody‑specific structures and delivers a dedicated toolkit for screening, redesign and de novo generation of high‑fitness binders.
🔨 Applications:
Zero‑shot NGS enrichment – ranks next‑generation sequencing hits by structural perplexity, boosting binder prevalence above random in 42/44 IL‑6 or SARS‑CoV‑2 mutant libraries (e.g. PMS variant: 0.82 vs 0.54 baseline).
Few‑shot binder (re)design – generates full‑VHH or CDR‑H3 variants conditioned on a single template; nanoFOLD sequences achieve the highest AUC with simple CNN oracles on both IL‑6 and SARS‑CoV‑2.
Nanobody scaffold optimisation – outperforms general (ESM‑IF) and antibody‑specific (AntiFold) models in sequence recovery, enabling rapid stability/affinity engineering without experimental structures.
📌 Key Insights:
Multiple data regimes – Three variants (xal, model, ft) reveal complementary strengths, with some models excelling on experimentally solved structures while others generalise best to large in‑silico sets.
Fitness‑landscape signal – nanoFOLD‑ft enriches binders in >90 % of SARS‑CoV‑2 mutants and >93 % of IL‑6 mutants, despite not seeing antigen context.
Generative robustness – oracle evaluation shows nanoFOLD variants preserve desired binding phenotype more reliably than baselines, especially when redesigning only CDR‑H3, underscoring its structure‑aware priors.
PoseX: AI Docking Tops Physics Rules for Real‑World Protein‑Ligand Cross‑Docking 🧪
Traditional physics‑based docking has long been the yard‑stick for predicting how drug‑like molecules bind to proteins, yet it struggles with receptor flexibility and scale. PoseX introduces the first comprehensive, training‑free benchmark that pits 22 modern algorithms—including five classical physics engines, 11 AI docking models and six AI co‑folding foundation models—against 718 self‑docking and 1 312 cross‑docking tasks derived from PDB structures published after 1 Jan 2022 (minimising information leakage). A lightweight OpenMM‑based relaxation pipeline and a public leaderboard complete the suite, allowing researchers to test a new method in minutes and see where it truly stands.
🔨 Applications:
Rapid method benchmarking – one command downloads the dataset, runs evaluation scripts and posts to a live leaderboard, accelerating algorithm iteration.
Generalisation stress‑tests – cross‑docking split lets pharma teams measure how a model copes with unseen pockets, a real bottleneck in lead‑optimisation.
📌 Key Insights:
AI wins—decisively. On the gold‑standard Astex set, SurfDock and Uni‑Mol hit 94 % pose success (≤2 Å) vs ~65 % for Glide/Vina; on PoseX cross‑docking, SurfDock still scores 75 %, dwarfing physics engines (<32 %).
Pocket‑aware beats pocket‑agnostic. Adding an explicit binding‑site mask (DiffDock‑Pocket) lifts cross‑dock success +5 pp over vanilla DiffDock, proving receptor context still matters in generative models.
Co‑folding FMs plateau. AlphaFold 3, Protenix and Chai‑1 reach ~60 % but stall on flexible targets, hinting at a need for pose‑specific fine‑tuning rather than sheer model scale.
Robustness tracks pocket similarity. For most AI models, RMSD grows sharply once TM‑score < 0.7; SurfDock and Uni‑Mol degrade least, signalling better learned inductive biases.
Did you find this newsletter insightful? Share it with a colleague!
Subscribe Now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website