AI in Life Science: Weekly Insights
Weekly Insights | April 27, 2025
In this issue:
Welcome back to your weekly dose of AI news for Life Science!
This week, we have some exciting new models lined up for you:
C2S-Scale: Scaling Large Language Models for Next-Generation Single-Cell Analysis 🧬
PepINVENT: Generative Peptide Design Beyond Natural Amino Acids 🧪
BoltzDesign1: Inverting Atomic AI for Generalized Biomolecular Binder Design 🧪
Dive into these game-changing innovations and explore how they are transforming the biotech and healthcare landscapes!
C2S-Scale: Scaling Large Language Models for Next-Generation Single-Cell Analysis 🧬
Current single-cell foundation models (scFMs) face limitations in scalability, flexibility across tasks, and integration of textual data, hindering their ability to synthesize insights across datasets and modalities. Introducing C2S-Scale leverages large language models (LLMs) trained on a multimodal corpus of over 1 billion tokens, including 50 million single-cell transcriptomes, biological text, and metadata. By converting scRNA-seq profiles into rank-ordered "cell sentences," C2S-Scale bridges transcriptomic data with natural language, enabling advanced biological reasoning and generative tasks.
🔨 Applications:
Perturbation Response Prediction: Accurately predicts transcriptional responses to unseen combinatorial perturbations (e.g., drug treatments, cytokine stimulation).
Natural Language Interpretation: Generates cell-type annotations, cluster captions, and dataset summaries in natural language.
Virtual Cell Platforms: Supports large-scale in silico experiments for hypothesis generation and therapeutic discovery.
📌 Key Insights:
Scaling Laws: Model performance improves consistently with size (410M to 27B parameters), data volume, and context length (up to 8k tokens).
Multimodal Integration: Combines transcriptomic data with biological text (e.g., paper abstracts) for richer representations.
Reinforcement Learning: Group Relative Policy Optimization (GRPO) enhances task-specific alignment (e.g., +15% accuracy in perturbation prediction).
Novel Metrics: Introduces scFID, a single-cell Fréchet Inception Distance, to evaluate generative models biologically.
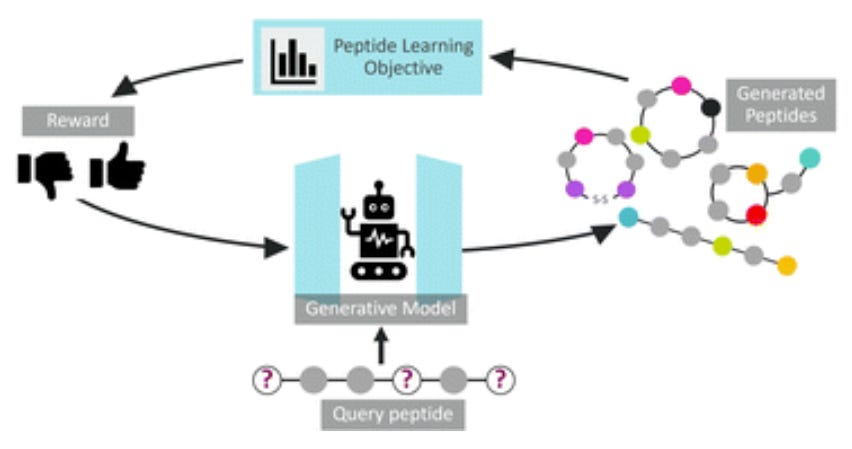
PepINVENT: Generative Peptide Design Beyond Natural Amino Acids 🧪
Traditional peptide design tools are constrained to natural amino acids, limiting exploration of the vast chemical space needed for optimizing therapeutic properties like stability, permeability, and target specificity. PepINVENT addresses this by integrating non-natural amino acids (NNAAs) through a generative AI framework, enabling de novo peptide design with enhanced flexibility and functionality.
🔨 Applications:
Multi-Parameter Optimization (MPO): Balances solubility, permeability, and bioactivity (e.g., enhancing HIV Rev-binding peptide permeability by 30%).
Topology-Specific Design: Generates linear, cyclic (head-to-tail, sidechain-to-tail), and disulfide-bridged peptides.
Lead Optimization: Modifies pharmacophores while preserving bioactivity (e.g., replacing alanines in macrocyclic peptides).
Peptidomimetics: Designs novel scaffolds mimicking protein interactions for undruggable targets.
📌 Key Insights:
CHUCKLES Representation: Atomic-level SMILES encoding ensures valid, diverse peptide structures (99% validity, 70× greater amino acid diversity compared to natural-only models) while capturing stereochemistry and backbone modifications.
Reinforcement Learning (RL): Group Relative Policy Optimization (GRPO) steers generation toward desired properties (e.g., macrocycles with >90% validity).
Novel Chemical Space: Generates 92,000+ novel NNAAs, expanding beyond the training set’s 10,000 NNAAs.
BoltzDesign1: Inverting Atomic AI for Generalized Biomolecular Binder Design 🧪
Existing protein binder design tools struggle with computational inefficiency, limited target diversity, and rigid ligand modeling, restricting their utility for complex biomolecular systems. BoltzDesign1 reimagines all-atom structure prediction models (Boltz-1/AlphaFold3) by inverting their architecture to enable zero-shot design of high-affinity binders for proteins, small molecules, nucleic acids, and post-translationally modified targets. By optimizing the atomic distance probability distribution, BoltzDesign1 reduces computational costs by 80% while achieving state-of-the-art in silico success rates.
🔨 Applications:
Multispecific Binder Design: Designs protein binders for metals (Zn²⁺, Fe³⁺), DNA/RNA, and post-translational modifications (phosphorylation, glycosylation).
Flexible Ligand Modelling: Predicts dynamic ligand conformations during binding, unlike fixed-backbone approaches.
📌 Key Insights:
High Success Rates: Achieves 85% in silico success for small-molecule binders vs. 50% for RfDiffusionAA.
Structural Diversity: Generates 2.5× more diverse backbones (average TM-score = 0.36 vs. 0.46) with tunable secondary structures via helix loss.
Cross-Model Consistency: Designs show <2 Å RMSD between Boltz-1 and AlphaFold3 predictions, validating robustness.
Efficient & Fast: Directly shapes atomic distance distributions via Pairformer/Confidence modules, bypassing 200-step diffusion backpropagation (3× faster than RfDiffusionAA).
Did you find this newsletter insightful? Share it with a colleague!
Subscribe Now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website