AI in Life Science: Weekly Insights
Weekly Insights | November 6, 2024
In this issue:
Welcome back to your weekly dose of AI news for Life Science!
This week, we have some exciting new models lined up for you:
MetaDegron: Multimodal protein language model for predicting E3 ligase interactions (degrons) 🪢
AlabOS: Reconfigurable workflow for autonomous laboratories🧫
Dive into these game-changing innovations and explore how they are transforming the biotech and healthcare landscapes!
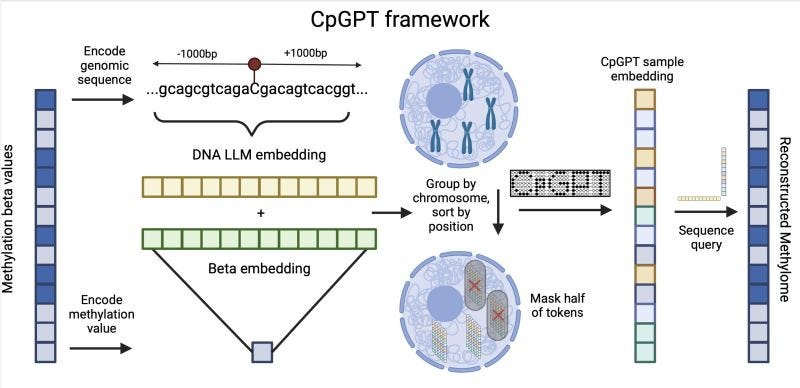
CpGPT: New foundation model for DNA methylation 🧬
DNA methylation is key for gene expression and chromatin structuring, with CpG sites often varying in diseases like cancer and aging. While important, most genomic foundation models do not capture well epigenetic and methylation signature. Introducing CpGPT, the latest foundation model specific for DNA methylation (code available soon at pyaging)!
📌 Key Insights:
Trained on over 100K samples from 1,500+ studies, covering 1M+ CpG sites.
CpGPT imputes methylation from minimal data, mapping different Illumina arrays to a common reference.
Fine-tuned CpGPT ranks 2nd overall and 1st on the public leaderboard for age and mortality prediction in the Biomarkers of Aging Challenge.
When fine-tuned, CpGPT accurately imputes methylation in new mammalian species.
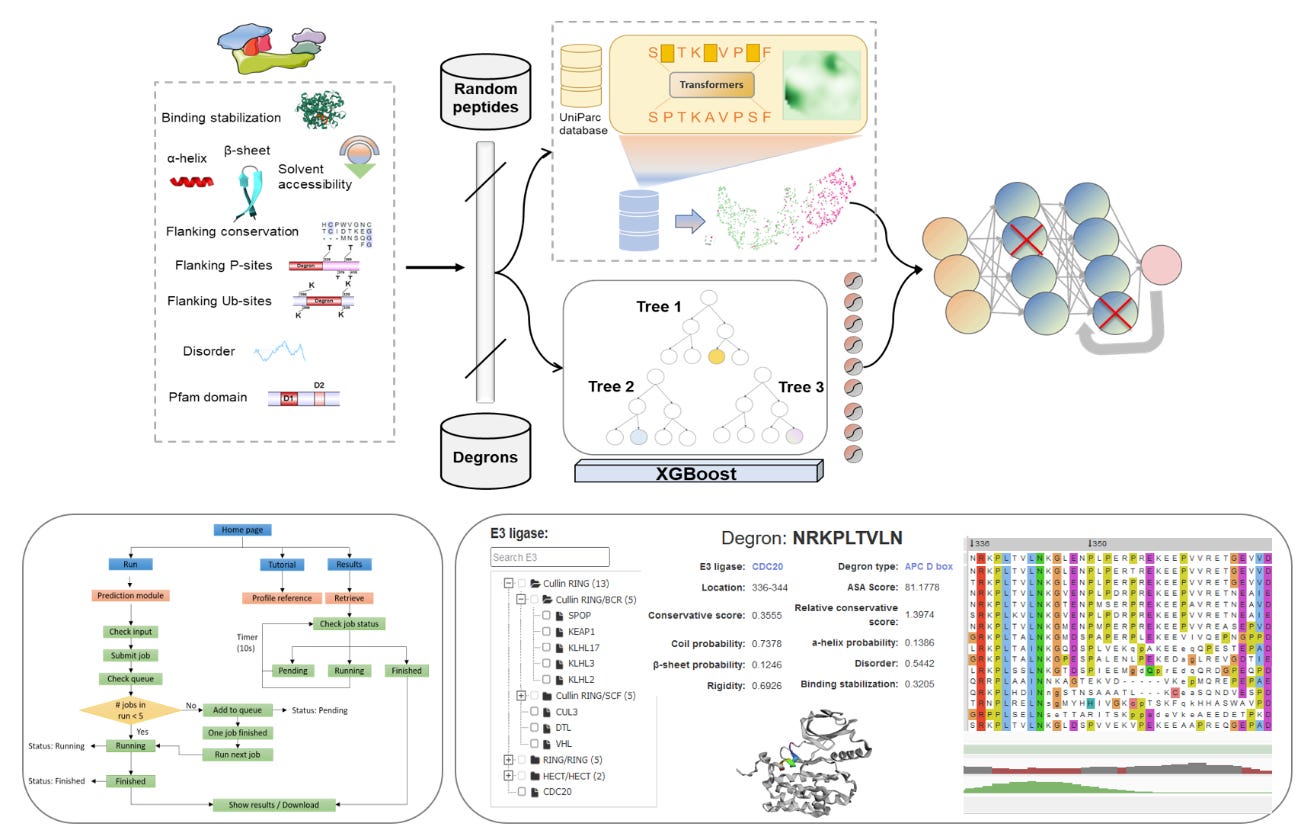
MetaDegron: Multimodal protein language model for predicting E3 ligase interactions (degrons) 🪢
Protein degradation through the ubiquitin proteasome system at the spatial and temporal regulation is essential for many cellular processes. E3 ligases and degradation signals (degrons), the sequences they recognize in the target proteins, are key parts of the ubiquitin-mediated proteolysis, and their interactions determine the degradation specificity and maintain cellular homeostasis. To date, only a limited number of targeted degron instances have been identified out of 600 existing. Introducing MetaDegron, a deep-learning models for predicting E3 ligase targeted degron!
📌 Key Insights:
Multi-modal protein language model combining combines contextual information, sequence-based features, and structural features to improve prediction performance.
Trained on E3 binding consensus patterns from the ELM database and a curated dataset of 300 degron instances from previous publications.
It enabled prediction of targeted degrons of 21 E3 ligases, and provides functional annotations and visualisation of multiple degron-related structural and physicochemical features.
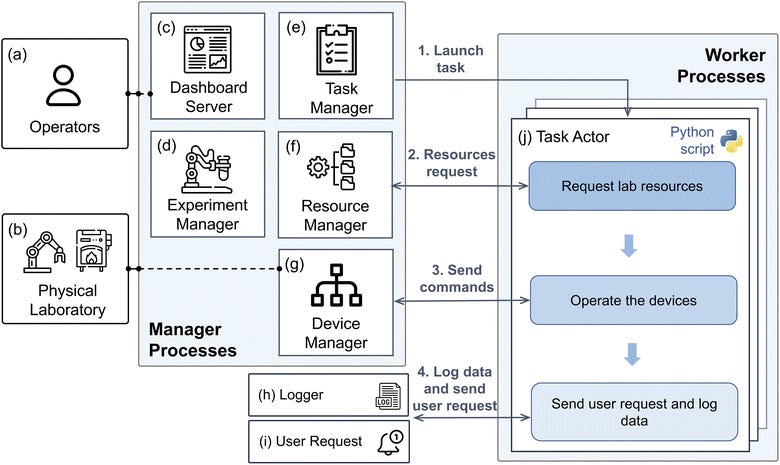
AlabOS: Reconfigurable workflow for autonomous laboratories🧫
The increasing complexity of autonomous laboratories in science requires sophisticated workflow management to efficiently handle the diverse tasks involved. Introducing AlabOS, a flexible software framework designed to orchestrate and simplify experiments and manage resources efficiently!
📌 Key Insights:
A platform-independent framework that’s user-friendly, requiring only basic knowledge of databases for adoption.
Provide solutions for autonomous laboratory workflow management, avoiding conflicts between tasks running at the same time in the laboratory.
It is already deployed in the A-Lab, an autonomous laboratory for inorganic materials synthesis housed at Lawrence Berkeley National Laboratory (LBNL).
Enables streamlined synthesis and characterisation processes, already supporting the creation of over 3,500 unique samples.
Did you find this newsletter insightful? Share it with a colleague!
Subscribe Now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website