AI in Life Science: Weekly Insights
Weekly Insights | July 29, 2025
In this issue:
Welcome back to your weekly round-up of new tools and methods in life sciences research.
This week, we’re spotlighting three innovations pushing the limits of segmentation, molecular representation, and MRI generalization:
MRI-CORE: A Generalist MRI Foundation Model Built from 110K Scans 🧠
PRISME: Unified Gene Embeddings from Sequence, Literature, and Graphs 🧬
Each of these tools helps researchers extract more insight from less data, whether that means fewer labels, broader coverage across imaging domains, or richer molecular signals across tasks.
What’s your most time consuming task along the drug discovery process?
We will send you open-source tools specific to your pain point.
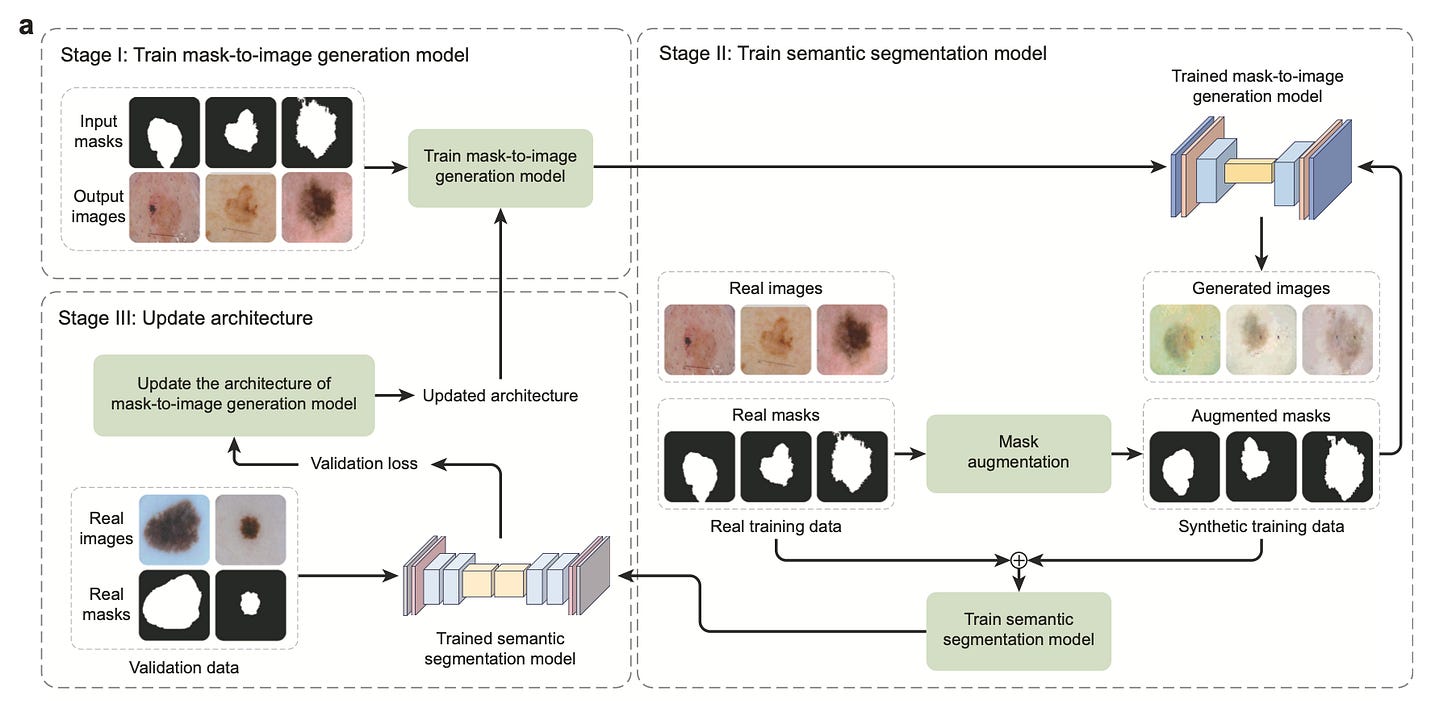
GenSeg: Better Segmentation with Just a Handful of Images 🧠
Training accurate segmentation models typically requires thousands of labeled images, which is not feasible in many clinical settings. GenSeg changes that by generating paired synthetic images and segmentation masks that directly improve segmentation performance in ultra-low data regimes. Rather than relying on augmentation or semi-supervision, it tightly couples a generative model with a segmentation objective in an end-to-end pipeline.
When tested on 11 segmentation tasks across 19 datasets, GenSeg enabled standard architectures like UNet and DeepLab to achieve up to 20% absolute improvement in Dice score with as few as 50 labeled samples. It also showed strong out-of-domain generalisation, requiring 8 to 20 times fewer labeled examples than traditional training approaches.
🔬 Applications
Low-Label Training - Enables high-quality segmentation with as few as 9 to 50 labeled images, reducing annotation requirements by up to 90 percent compared to standard methods.
Architecture Compatibility - Improves segmentation accuracy for UNet, DeepLabv3+, and other models without changes to architecture or training routine.
Multi-Domain Transfer - Achieves performance gains across 11 tasks and 19 datasets including breast, colon, gland, and polyp segmentation.
📌 Key Insights
Up to 20 point Dice improvement - Models trained with GenSeg on just 50 labeled samples improved Dice scores by 8 to 20 points over baseline training.
Matches full supervision with 10× less data - In foot ulcer and polyp segmentation, GenSeg matched fully supervised performance using only 10 percent of the labels.
Cross-dataset generalisation - Improved out-of-domain accuracy by 10 to 15 points when evaluated on unseen datasets with different staining or imaging modalities.
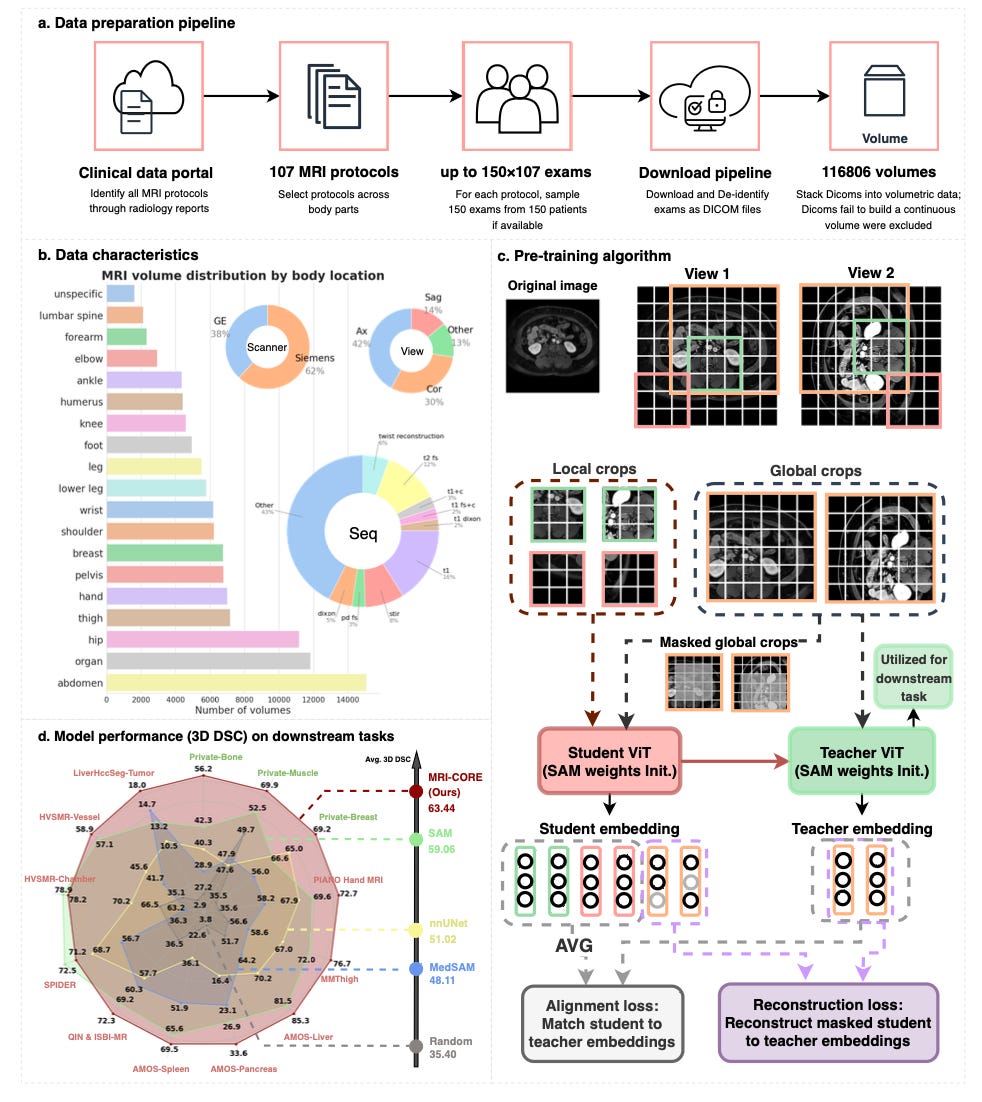
MRI-CORE: A Foundation Model for MRI That Actually Transfers 🧪
MRI-CORE is a vision foundation model trained on more than 6 million slices from 110,000 MRI volumes, covering 18 body regions. Built to address the lack of generalisable MRI models, it delivers strong performance on segmentation, classification, and zero-shot tasks with minimal labeled data. It also comes with a novel benchmarking setup showing how pretraining similarity affects downstream results.
Across 11 datasets, MRI-CORE outperformed nnU-Net by up to 33.3% and SAM by up to 17.4% in 3D Dice scores. In zero-shot settings, it captured sharper boundaries than other foundation models, and its image representations outperformed SAM and MedSAM in predicting anatomical regions, acquisition parameters, and even patient sex and disease severity.
🔨Applications
Few-Shot Segmentation - Outperforms task-specific baselines using as few as 5 training examples, with Dice scores exceeding 70 percent on liver, spleen, and hand.
Zero-Shot Label-Free Segmentation - Achieves 45 to 60 percent Dice in several tasks without any labeled training data by clustering pixel embeddings.
Embedding-Based Metadata Prediction - Predicts imaging institution, acquisition plane, scanner type, and anatomical region with over 85 percent top-1 accuracy.
📌 Key Insights
33.3 percent gain over nnU-Net - Achieved the highest 3D Dice score on Private-Bone (81.2 percent) compared to 60.9 percent with nnU-Net.
17.4 percent gain over SAM - Surpassed SAM on Private-Muscle by 17.4 points in 3D segmentation.
Top classification accuracy - Embeddings correctly predicted imaging site in 99.7 percent of cases and sex in 93.2 percent, outperforming MedSAM and SAM.
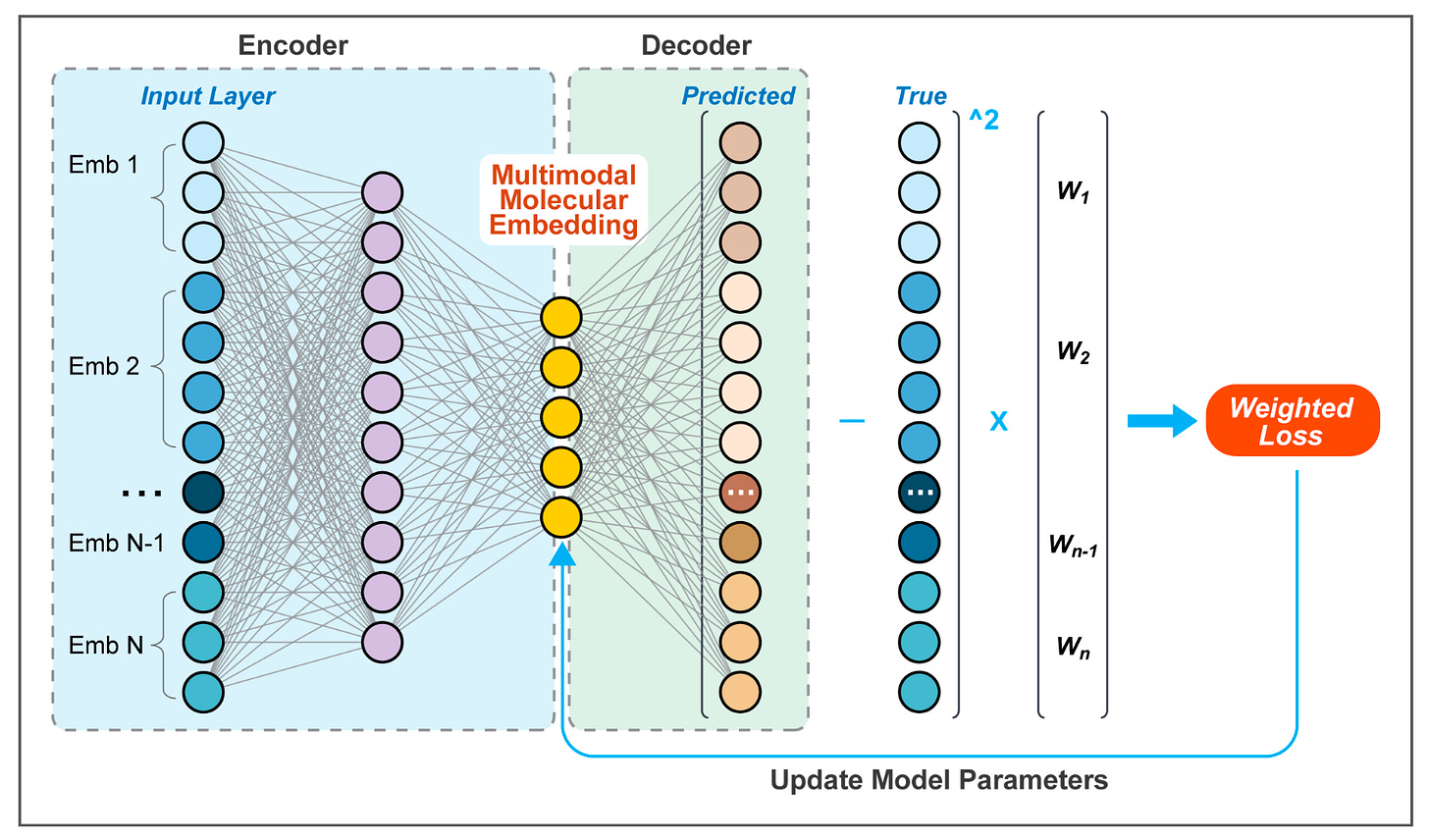
PRISME: A Multimodal Platform for Gene Embedding Integration 🧬
Different gene embeddings capture different types of biological signal, but most downstream tasks require a broader view. PRISME solves this by integrating multiple molecular embeddings—sequence-based, literature-derived, and graph-based, into a single, low-dimensional representation using an autoencoder. This unified embedding outperforms or matches the best individual methods across a range of biomedical tasks.
On benchmark tasks, PRISME achieved the highest accuracy (77%) and AUC (0.85) for gene-gene interaction prediction, and showed strong performance in missing value imputation, recovering signal from incomplete data. It also provides tools for assessing embedding complementarity using an adjusted SVCCA metric.
🔨Applications
Cross-Task Generalisation - Improves predictive accuracy in gene–gene interaction, protein–protein interaction, gene–disease association, and subcellular localisation.
Robust to Missing Embeddings - Maintains strong performance in downstream tasks even when only a subset of input embeddings is available.
Unsupervised Representation Learning - Outperforms single-source embeddings without needing task-specific retraining.
📌Key Insights
Gene-gene interaction - Achieved 0.77 accuracy and 0.85 AUC, outperforming all single-embedding baselines.
Protein–protein interaction - Reached 0.76 accuracy and 0.83 AUC, surpassing deepwalk and node2vec by over 5 points.
Embedding complementarity - Adjusted SVCCA revealed low redundancy between modalities, validating that multi-source fusion improved task-specific accuracy by 3 to 9 points.
Did you find this newsletter insightful? Share it with a colleague!
Subscribe Now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website