AI in Life Science: Your Weekly Insights
Welcome back to your weekly dose of AI news for Life Science!
What if we could design molecules with unprecedented precision, fuel AI with comprehensive biological datasets, and simulate complex disease mechanisms virtually, all before stepping into the wet lab?
This week's breakthroughs bring us closer to that reality, fundamentally reshaping how we approach biological research and accelerate discovery.
Reminder: Our Tools Atlas database contains the code to all open-source tools mentioned in this issue and all previous newsletters!
What’s your most time consuming task along the drug discovery process?
We will send you open-source tools specific to your pain point.
ProDomino: Engineering Molecular Precision, No More Guesswork 🧬
💡The Breakthrough: No More Guesswork in Protein Engineering!
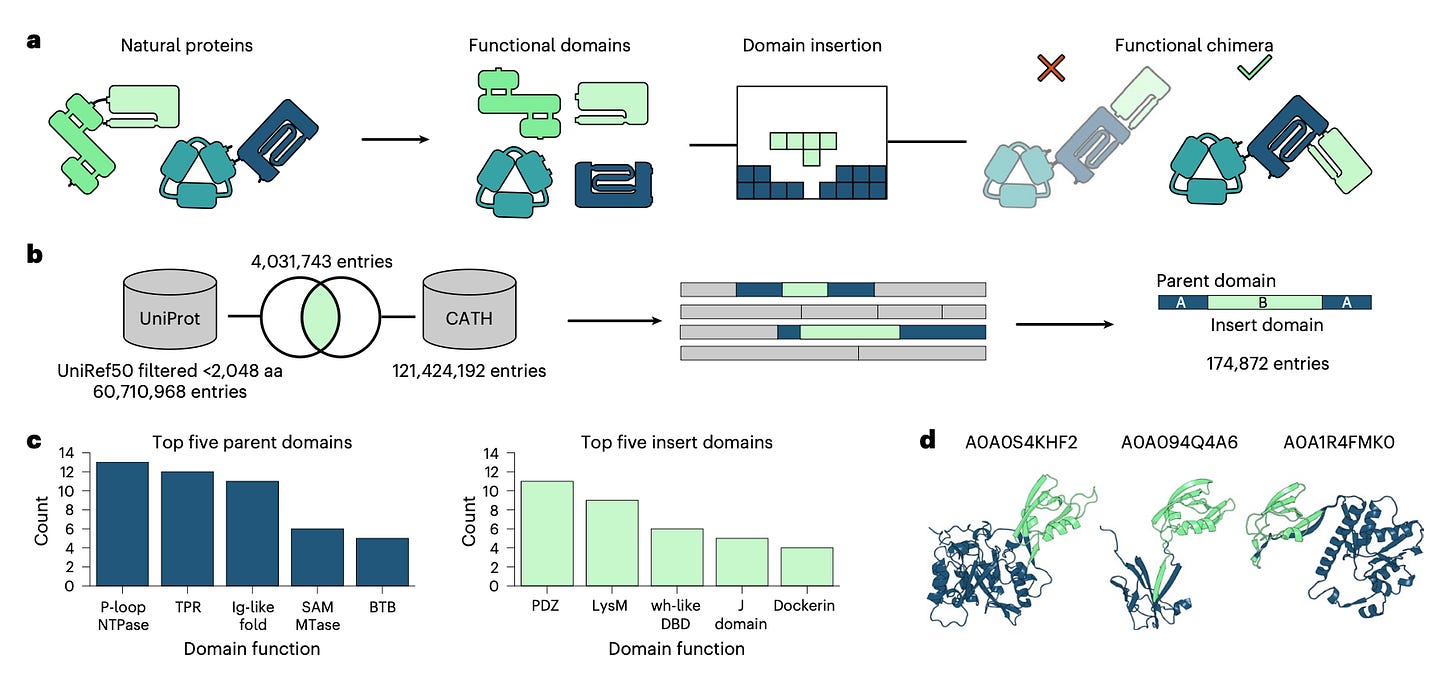
Designing proteins with specific, controllable functions, like turning a gene editor on or off with a flash of light, has long been a labor-intensive endeavor. ProDomino is fundamentally changing the game. This AI platform acts as a molecular architect, precisely predicting optimal sites for domain insertions, transforming the arduous process of creating allosteric switches into a rational, one-shot design sprint.
🤿 Diving Deeper Into How it Works: Learning from Nature's Blueprints
ProDomino's secret lies in its training data, which goes beyond scarce experimental results. Researchers curated a massive dataset of 174,872 protein sequences from "naturally occurring intradomain insertion events". Essentially, nature's own successful protein fusions. By learning from millions of evolutionary successes, ProDomino accurately predicts where new domains can be inserted without breaking the protein. This means less time in the lab and more reliable designs!
📈 Transformative Uses & What This Means For Your Research:
One-Shot Design: Rapidly creates potent, single-component protein switches, accelerating the design of custom allosteric proteins for diverse applications.
Precision Genome Editing: Used to design novel light- and chemically-regulated variants of CRISPR-Cas9 and Cas12a nucleases, offering unprecedented spatiotemporal control over gene editing. This significantly enhances safety and efficacy in gene therapy by allowing precise control over when and where edits occur.
📌 The Metrics That Matter:
High Success Rates: The pipeline's predictions were experimentally validated in both E. coli and human cells, achieving success rates of approximately 80%. Overall, experiments confirmed the model's predictions in 78% of cases.
Predictive Power in Practice: For an antibiotic resistance enzyme, a CAT-LOV2 hybrid created with the platform showed a 20-fold difference in optical density between light and dark conditions. For a Cas12a variant, the model successfully predicted an allosteric site that resulted in a threefold reduction in activity under light conditions.
Broad Applicability: ProDomino's predictive performance on the AraC bacterial transcription factor had an AUROC of 0.84, indicating its strong ability to generalize and correctly identify allosteric sites, even in complex, multi-domain proteins like CRISPR-Cas nucleases.
The Peptide Powerhouse: Fueling AI with Foundational Data 📊
💡The Breakthrough: A New Era for Peptide Therapeutics Data!
AI thrives on data, but the peptide therapeutics field has been starving. Current databases often lack crucial information on multi-functional peptides and high-quality structural data, creating a bottleneck for next-generation AI. Now, a new foundational dataset has arrived, providing the much-needed fuel for AI-driven peptide discovery and accelerating our understanding of these versatile molecules.
🤿 Diving Deeper Into How it Works: Building the Google Maps of Peptides
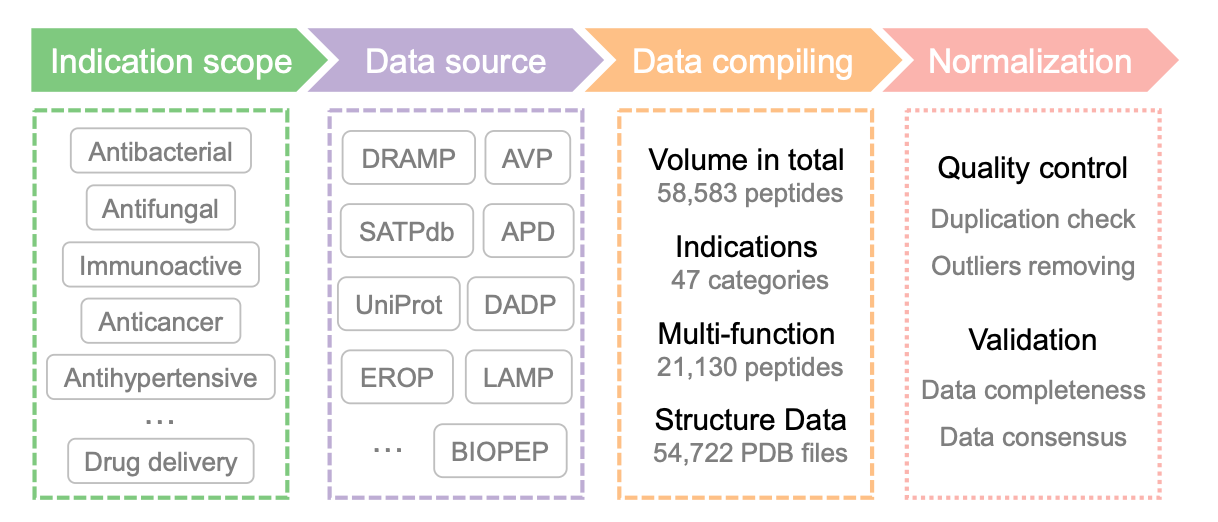
This new resource is a monumental leap, containing 58,583 experimentally validated therapeutic peptides. Crucially, it specifically addresses past limitations by focusing on:
Multifunctionality: It boasts 21,130 multifunctional peptides, more than double the 9,986 entries in the previous largest database. This is vital for understanding how peptides exhibit "moonlighting characteristics" and for drug repurposing.
Structural Richness: It provides high-quality structural annotations for 54,722 peptides, a massive increase from just 16,131 entries previously. This was made possible by integrating cutting-edge tools like AlphaFold2, ensuring the data is ready for structure-aware AI models.
📈 Transformative Uses & What This Means For Your Research:
Accelerating Computational Discovery: This dataset provides the necessary high-quality data to build and train computational pipelines for therapeutic peptide discovery, including predicting antimicrobial peptides and repurposing existing peptide drugs.
Unlocking New Insights: The resource facilitates a deeper exploration of the fundamental "sequence-structure-function" relationships that are essential for understanding the underlying principles of peptide drug design.
📌 The Metrics That Matter:
Unprecedented Scale: With 58,583 peptides across 47 functional categories, the dataset is the most comprehensive of its kind, far surpassing the size and scope of previous efforts.
Doubling Multifunctionality: Features 21,130 multifunctional peptides, significantly advancing research into peptides with diverse therapeutic potentials.
Massive Structural Upgrade: Provides 54,722 structurally annotated peptides, a nearly 3.4x increase over prior efforts, powering the next wave of structure-informed AI.
MintFlow: Your Virtual Lab for Tissue Simulation 🔬
💡The Breakthrough: Simulating Tissue Behavior, Not Just Describing It!
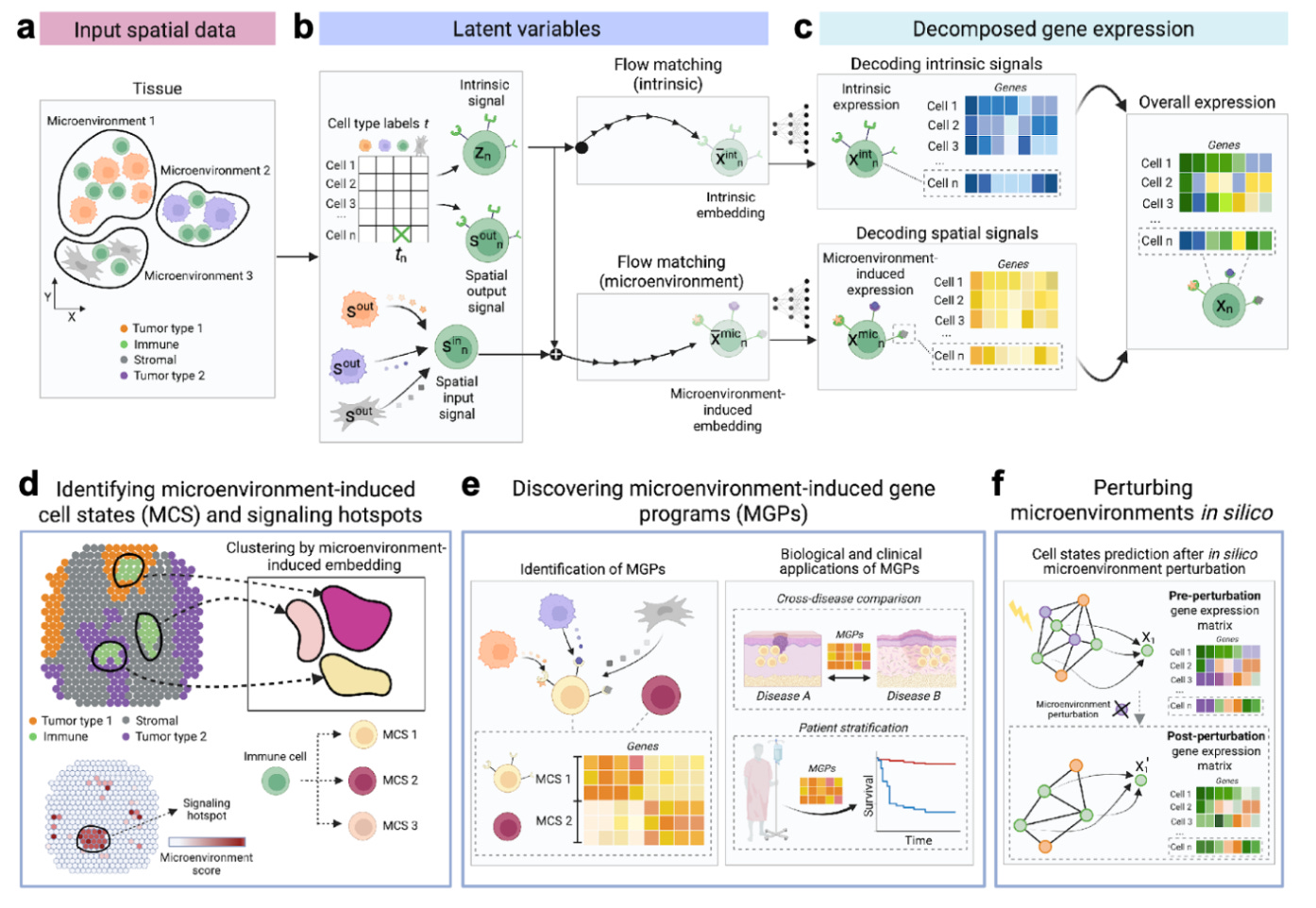
Spatial transcriptomics gives us stunning maps of cells in tissues, but traditional analysis stops at description. MintFlow transcends this, offering a generative AI framework that can predict how cells behave and respond to changes in their microenvironment. It's like having a virtual lab where you can test biological hypotheses that are simply "unfeasible" to perform in the physical world.
🤿 Diving Deeper Into How it Works: Predicting Cause and Effect in Tissues
MintFlow's core innovation is its ability to separate a cell's gene expression into its inherent identity ("intrinsic variability") and the influence of its neighbors ("microenvironment-induced" expression). By modeling cellular communication, it can perform "in silico perturbations"—virtual experiments where you can, for example, "delete or replace" certain cells and predict the ripple effects on gene expression across the tissue. This opens up entirely new avenues for understanding disease mechanisms and testing therapeutic strategies.
Case Study 1: Reversing Inflammation in Atopic Dermatitis: MintFlow accurately predicted that augmenting regulatory T cells (Tregs) would not promote inflammation, consistent with clinical data from patients responding positively to dupilumab, who showed increased skin Tregs.
Case Study 2: Targeting Immunosuppression in Kidney Cancer: In a simulation for clear cell renal cell carcinoma (ccRCC), MintFlow identified an "immunosuppressed border population" of T cells within tumor lymphoid structures. Critically, an in silico "macrophage depletion" predicted a "reversion" of these T cell states towards a more functional phenotype, directly linking a cause (macrophage interaction) to a potential therapeutic effect..
📈 Transformative Uses & What This Means For Your Research:
Mechanistic Discovery: Uncovers spatially encoded drivers of disease, identifying fine-grained cellular states, like immunosuppressed T cells in kidney cancer, that traditional methods miss.
Predictive Therapeutics: Simulates interventions, such as T cell replacement or spatially targeted macrophage depletion, to predict how they might reverse disease states and influence gene expression.
Patient Stratification: Directly connects in silico-predicted gene programs to real-world clinical outcomes, demonstrating a "survival advantage" for a reprogrammed T cell state in a large cohort of kidney cancer patients.
📌 The Metrics That Matter:
Predicting Clinical Outcomes: In kidney cancer, high expression of the original, immunosuppressed T cell program was significantly associated with worse overall survival (p=0.0073). Conversely, when the in silico "reprogrammed" T cell gene program was used, high expression was associated with a survival advantage (p=0.0034).
Simulating Therapeutic Effects: In atopic dermatitis, an in silico simulation of Treg augmentation was not associated with pro-inflammatory pathways. This aligned with clinical data showing that patients who responded well to treatment typically had an increase in skin Tregs.
Validated Perturbation: In a kidney cancer simulation, when immunosuppressed T cells were replaced with a signature resembling those post-ICB therapy, MintFlow accurately predicted the upregulation of 11 out of 12 "ground truth" macrophage genes known to be expressed after ICB treatment.
Did you find this newsletter insightful? Share it with a colleague!
Subscribe Now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website