🧠 AOP Net: Mapping the Hidden Logic of Toxicity
Deep Dive | Edition 6
Welcome back to the deep dive, where we break down the AI tools and data reshaping how new drugs are discovered. In each edition, we speak directly with the teams behind these tools to explain what they solve, how they work and where they are going next.
Today, we’re diving into AOP Net, an AI-enhanced platform built by Abhik Seal, a researcher at Indiana University Bloomington. With a background in cheminformatics and systems pharmacology, Abhik has long been interested in how network analysis and AI can uncover the hidden logic of toxicity.
What began as a side project to organise complex toxicological data has now grown into a powerful new tool for understanding how chemicals cause harm, and for rethinking the future of safety science. An exciting development for a field that remains surprisingly underexplored.
🔴 The Problem
The Adverse Outcome Pathway (AOP) framework was designed to help scientists map how chemical exposures cause harm. Each AOP starts with a molecular initiating event and ends with an adverse outcome like liver fibrosis, neurodegeneration, or reproductive failure. In theory, this helps researchers and regulators trace cause and effect across biological systems.
In practice, AOPs are messy. Many are incomplete, missing connections between events, or structured as linear chains that ignore biological complexity.
As Abhik put it, “Some AOP graphs are incomplete. Nodes lack edges, and there’s incorrect or jumbled information, particularly regarding cause-and-effect relationships.”
That becomes a major bottleneck when researchers want to explore how different compounds affect overlapping pathways, or when regulators need to understand toxicity beyond a single mode of action.
Beneath that complexity, however, sits a surprising pattern. Many drug-induced toxicities are not unique. They trace back to a handful of tightly connected failure points, especially mitochondrial dysfunction, oxidative stress, and reactive oxygen species. These core mechanisms appear again and again, but have rarely been studied together as a system.
💡 The Idea
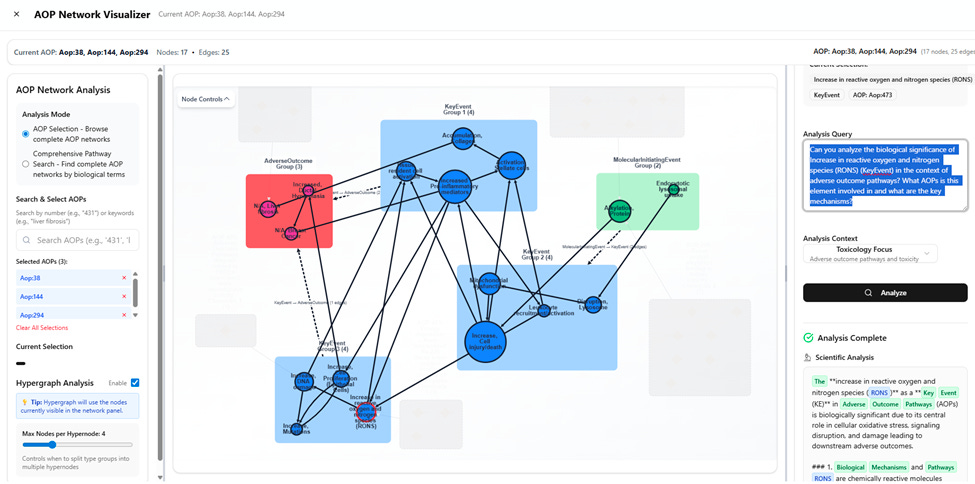
AOP Net imports the full AOP-Wiki database and renders it as a live, interactive network. Users can zoom in on specific pathways, search across nodes, filter by evidence strength, and visualise how different adverse outcomes are mechanistically connected.
But the real shift comes from its AI analysis panel, which combines the structure of the AOP network with a large language model. When a user selects a node, such as oxidative stress, the AI explains its biological significance, connected diseases, and regulatory relevance in real time.
It supports multiple analysis modes, including drug, disease, mechanism, regulatory, and pathway. Each mode applies a different reasoning style, allowing scientists to ask different types of questions depending on their focus.
Behind the scenes, the system uses a three-layer prompting strategy to constrain the LLM and keep its output grounded in the selected biology. It injects context from the graph and formats responses based on the chosen analysis style.
The platform also runs real-time metrics on the network itself, calculating centrality scores and identifying convergence points. Oxidative stress, for example, emerged as the most connected event in the network: linked to 23 upstream pathways and 56 downstream effects.
📊 What about the data?
AOP Net draws directly from the public AOP-Wiki, which contains:
232 Molecular Initiating Events
1,067 Key Events
200 Adverse Outcomes
Each node is annotated with ontology terms, stressors, and evidence levels. These are visualised in real time, and the network updates automatically when new AOPs are published.
In total, the system connects over 1,400 nodes with more than 2,100 relationships, which Abhik calls a “biological internet of toxicological responses.” What emerges from that network is a repeat pattern: oxidative stress, mitochondrial dysfunction, and ROS appear as highly connected hubs, not isolated events but core amplifiers of toxic response across pathways.
🔬 Why It’s Different
Most AI tools in toxicology focus on prediction, estimating toxicity, classifying risk, or flagging bad actors from structure. AOP Net is different. It works at the level of mechanism, helping scientists explore how biological damage unfolds across interconnected events. Pretty exciting stuff.
This shift creates several advantages:
● Mechanism-first insight: AOP Net does not just flag toxicity. It shows how a molecular event propagates toward disease through biological steps. It reveals how many toxicities trace back to the same few mechanistic bottlenecks.
● Context-aware analysis: The LLM interprets each event in context, using the surrounding network structure to refine its outputs.
● Multi-pathway comparison: Users can trace where different chemicals lead to the same endpoint or where one event diverges into multiple outcomes.
● Interactive and explainable: The system supports filters, layout algorithms, and reasoning modes so users can explore, question, and iterate, not just browse.
🔮 The Future
Today, AOP Net focuses on small molecules and environmental chemicals. RNA and gene therapy data are not yet integrated, though Abhik hopes to expand into those modalities.
One major feature in development is a curator mode, where the LLM can help users complete graphs by suggesting missing links, retrieving literature, and flagging inconsistencies.
Further out, Abhik envisions a role for AOP Net in personalised toxicology, adjusting pathways based on patient-specific factors like genotype, co-exposures, or disease state, but the infrastructure for that definitely still needs to mature. “The cost and logistical challenges of preparing individualised drugs, especially for general public access, are significant hurdles,” he noted.
For now, AOP Net is already moving the field forward. It gives researchers and regulators a tool to actively explore toxicological mechanisms, not just view them. And it shows how structured graph data and language models can work together: not to replace human reasoning, but to make it faster, deeper, and easier to access.
📄 Read the paper!
⚙️ Try it out on GitHub.
👨🔬 Get in touch with Abhik.
Thanks for reading!
Did you find this newsletter insightful? Share it with a colleague!
Subscribe now to stay at the forefront of AI in Life Science and keep up with this upcoming season of deep dives.
Connect With Us
Have questions or suggestions for our next deep dive? We’d love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website




Brilliant. I'm wondering how the platform infers those missing cause-and-effect relationships from such incomplete data? That's quite an algoritmhic challenge to solve.