Boltz's BLOSUM Is All You Learn, DeepMind's Cell2Sentence, and TU Wien's ChemTorch
Kiin Bio's Weekly Insights
Welcome back to your weekly dose of AI news for Life Science!
What’s your biggest time sink in the drug discovery process?
🧬 BLOSUM Is All You Learn: How Generative Models Echo Evolution
Even in the age of generative biology, evolution remains the best pretraining dataset there is.
Antibody design has become one of the most exciting frontiers in biological AI.
Diffusion models, inverse folding networks, and language models can now generate antibody sequences that actually bind, sometimes rivaling those evolved in nature.
But a simple question remained: what are these models really learning?
Researchers from Boltz and the University of Cambridge decided to find out.
Their paper, BLOSUM Is All You Learn, reveals that many state-of-the-art generative models are implicitly rediscovering evolutionary substitution patterns, the same amino acid rules encoded in classical BLOSUM matrices.
🔬 Applications and Insights
1️⃣ Evolutionary priors behind model scores
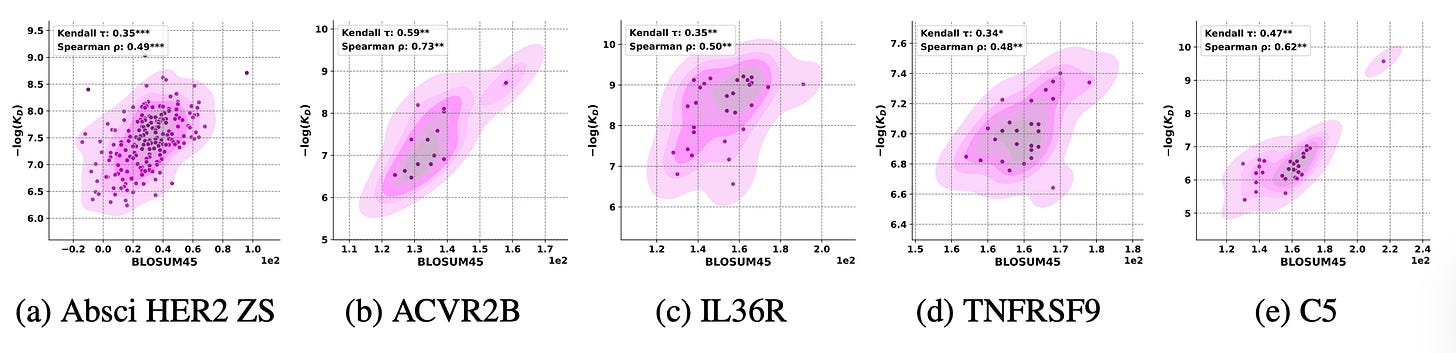
The team compared model log-likelihoods from DiffAbXL-A, AntiFold, and IgLM with traditional evolutionary similarity metrics like BLOSUM, PWM, and PSSM.
Across 14 antibody datasets, BLOSUM similarity to the parental sequence correlated just as strongly with binding affinity as the model scores themselves.
2️⃣ Models mirror evolutionary substitution patterns
DiffAbXL-A’s likelihoods showed Spearman correlations above 0.7 with BLOSUM similarity across multiple targets. The pattern held for AntiFold and IgLM too, proof that these models internalise evolutionary constraints without ever being told to.
3️⃣ BLOSUM outperforms modern metrics
When BLOSUM was computed against each antibody’s parent sequence, it matched or outperformed generative models at predicting affinity. Using global or consensus sequences broke the signal, showing that context matters and local evolutionary history still dominates.
4️⃣ Interpretable metrics still work
Simpler, interpretable measures like BLOSUM45 can rival deep models for antibody ranking, offering transparency and grounding in biochemical intuition.
💡 Why It’s Cool
This study turns a mirror on modern generative biology.
Despite their complexity, today’s antibody AI models seem to be relearning evolution, one substitution at a time.
It’s a humbling reminder: the statistical “priors” evolution built into life still form the foundation of model intelligence.
📄 Check out the paper!
⚙️ The code isn’t out yet!
🧫 Teaching AI to Read Cells: Google DeepMind’s Cell2Sentence Scale 27B
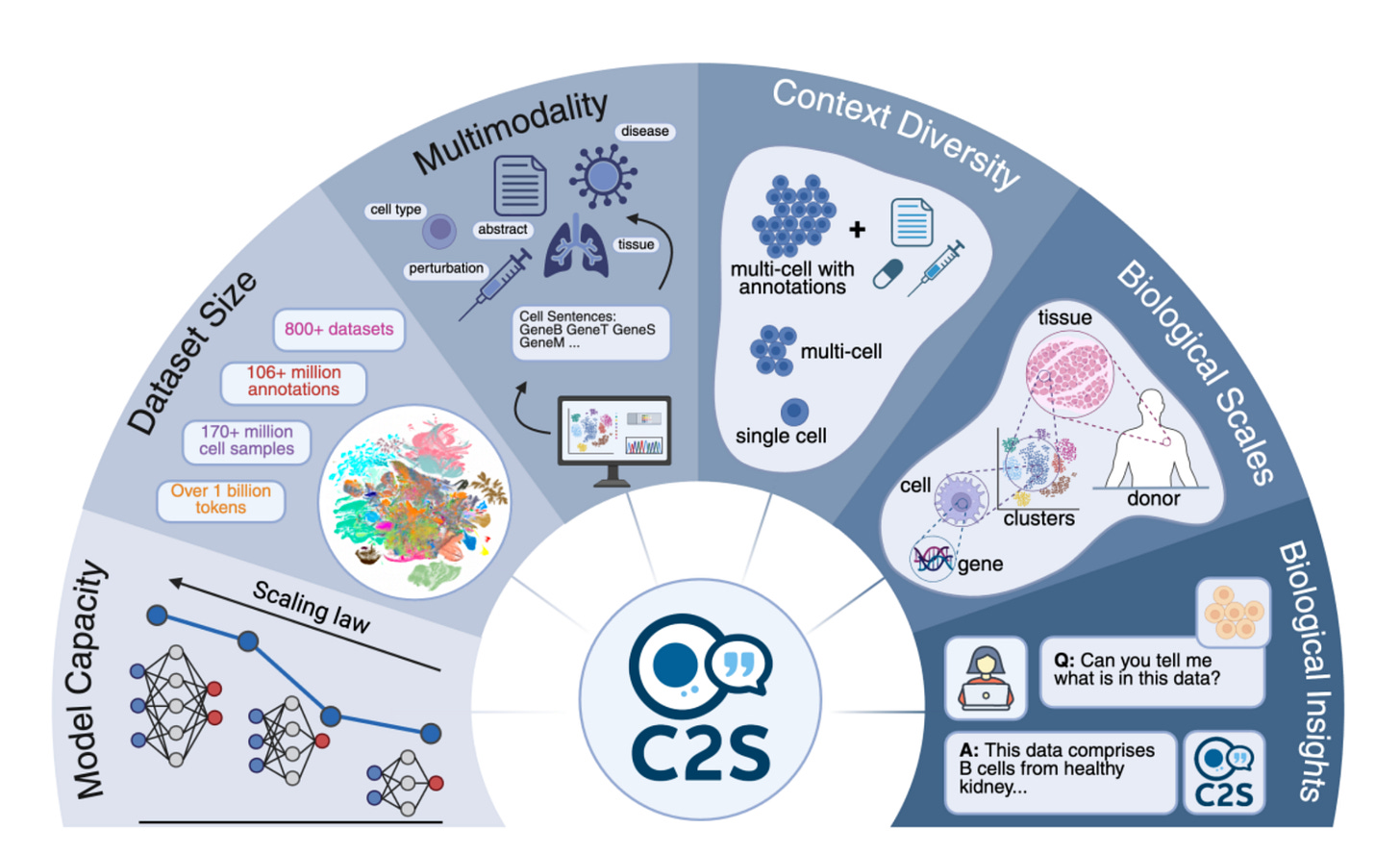
A collaboration between Google DeepMind and Yale University has produced Cell2Sentence Scale 27B (C2S-Scale 27B), a tool that lets AI and biology communicate, with huge potential to accelerate cancer research.
Built on DeepMind’s Gemma models, it can identify drugs that amplify immune signals. For cancer immunotherapy, this is transformative: “cold” tumours that hide from the immune system can be made “hot,” making them visible to immune attack and opening the door to more effective treatments.
By viewing cells as “sentences” and gene activity as a language AI can understand, C2S-Scale 27B opens new ways to explore cellular behaviour. For instance, it predicted that the kinase inhibitor silmitasertib (CX-4945) would increase antigen presentation in an “immune-context-positive” setting but have little effect in a neutral one.
This hadn’t been reported before, showing that the AI can generate new, testable hypotheses.
🔬 Applications and Insights
1️⃣ Interpreting Cellular Information
Each cell’s gene expression profile is transformed into a “sentence” of gene names, with the most active genes first. This encoding lets large language models interpret molecular data, effectively giving AI the ability to read the language of cells.
2️⃣ Predicting Cellular Responses
By learning how cells change gene expression in response to drugs, the model can simulate responses to new treatments, testing thousands of hypothetical drug-cell interactions far faster than traditional lab experiments.
3️⃣ Multimodal Reasoning
As the model has been trained on data from scientific papers, cell metadata, and biological annotations, C2S-Scale 27B can learn what the activity of certain genes signifies within the living system, capturing their function and significance.
4️⃣ Advancing Cancer Immunotherapy
Traditional single-cell analysis reveals tumour complexity but not therapeutic relevance. By interpreting cell data as “sentences,” the model can map how cells respond to signals, identify activation or suppression patterns, and link them to pathways and therapeutic targets.
Researchers can even prompt the AI with questions like, “Which genes predict a better T-cell response to checkpoint inhibitors?”, generating testable hypotheses about immune mechanisms.
💡 Why It’s Cool
As well as being an innovative way to analyse single-cell data, C2S-Scale 27B could be the start of a whole new language in biology, one that is comprehensible to LLMs.
By turning gene expression profiles into something these models can read, researchers can start to predict how cells will react to drugs, uncover patterns in tumours, and explore new therapies, all without a single wet lab experiment.
📄 Check out the paper!
⚙️ Try out the code.
⚗️ ChemTorch: Making Chemical Reaction Prediction Accessible to Everyone
What if predicting chemical reactions could be as simple as switching a few lines of code?
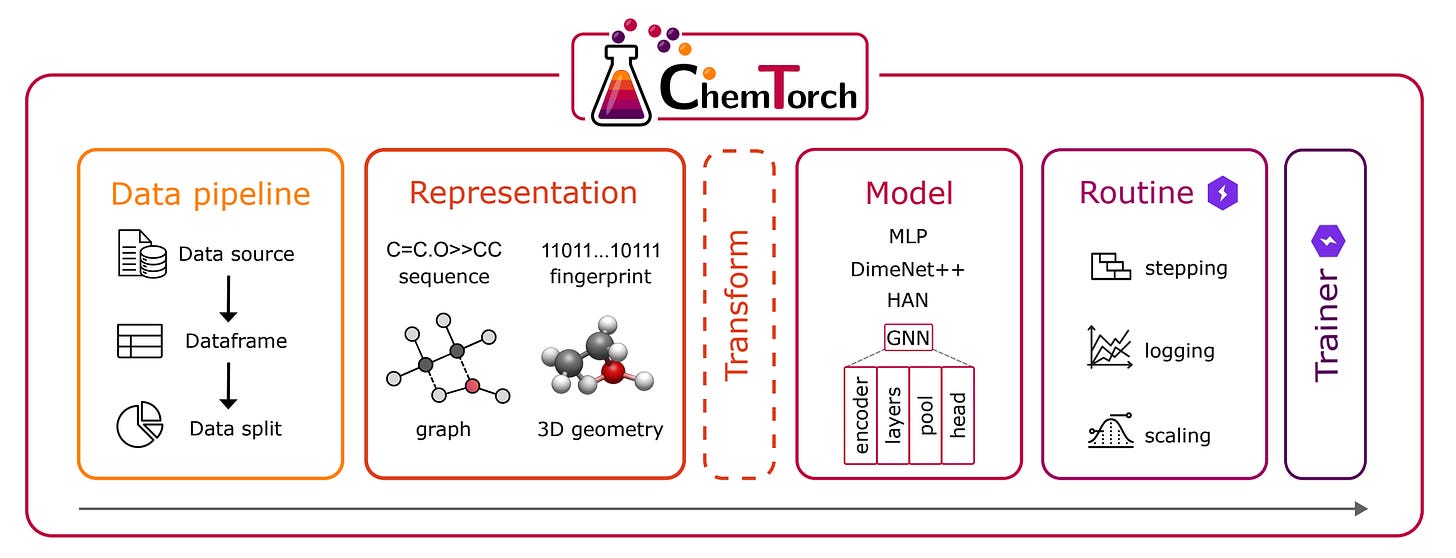
Researchers at TU Wien have just released ChemTorch, an open-source deep learning framework that’s set to transform how we model chemical reactions.
Instead of cobbling together scattered code from different repositories, scientists can now benchmark, compare, and develop reaction prediction models in one unified platform.
ChemTorch streamlines the entire workflow of building neural networks for chemistry, from data handling to model training to evaluation. Its modular design supports multiple reaction representations (fingerprints, SMILES sequences, molecular graphs, and 3D geometries) and architectures, all configurable without touching code.
🔬 Applications and Insights
1️⃣ Barrier Height Prediction
Benchmarking four approaches on 12,000 reactions showed graph-based models achieved 4.1 kcal/mol errors versus 14.5 kcal/mol for fingerprints. 3D geometry models reached 2.5 kcal/mol when transition states were available.
2️⃣ Data Efficiency
Structural models reached lower errors with just 1,000 training reactions than fingerprint models trained on the full dataset — crucial when reaction data is expensive to generate.
3️⃣ Out-of-Distribution Reality Check
Models that excelled on random test splits failed catastrophically when extrapolating to new reaction types or unseen barrier height ranges, highlighting why rigorous benchmarking matters.
4️⃣ Reproducibility by Default
Built-in data splitters, hyperparameter optimisation, and automatic logging mean every experiment can be exactly reproduced with a single command.
💡 Why It’s Cool
ChemTorch creates a foundation for community-driven progress by standardizing evaluation protocols and making diverse architectures directly comparable.
Whether you’re in synthesis planning, catalyst design, or high-throughput screening, it provides the infrastructure to move from concept to working model in hours, not weeks.
📄 Check out the paper!
⚙️ Try out the code.
Thanks for reading!
Did you find this newsletter insightful? Share it with a colleague!
Subscribe now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website