🧀 Deep MedChem: Screening Billions of Molecules in Seconds by Vectorizing 3D Shape and Electrostatics
Deep Dive | July 16, 2025
Welcome back to the Deep Dive series, where we break down the AI tools reshaping how new drugs are discovered. In each edition, we speak directly with the teams behind these tools to explain what they solve, how they work and where they are going next.
Today we look at Deep MedChem, the company behind CHEESE, a chemical‑embedding engine that shrinks tens of billions of purchasable molecules into a searchable vector index you can sweep in seconds. By converting full 3D shape and electrostatics into compact coordinates, CHEESE removes one of the heaviest bottlenecks in ligand based virtual screening.
This tool can be especially helpful for pre-screening and filtering (e.g. selecting a manageable subset of a large chemical space), hit expansion, building a Structure-Activity Relationship (SAR) by catalogue, diversity selection, property optimization, active learning, scaffold hopping or DEL screen processing.
What’s slowing you down? Let us tackle it.
🔴 The Bottleneck
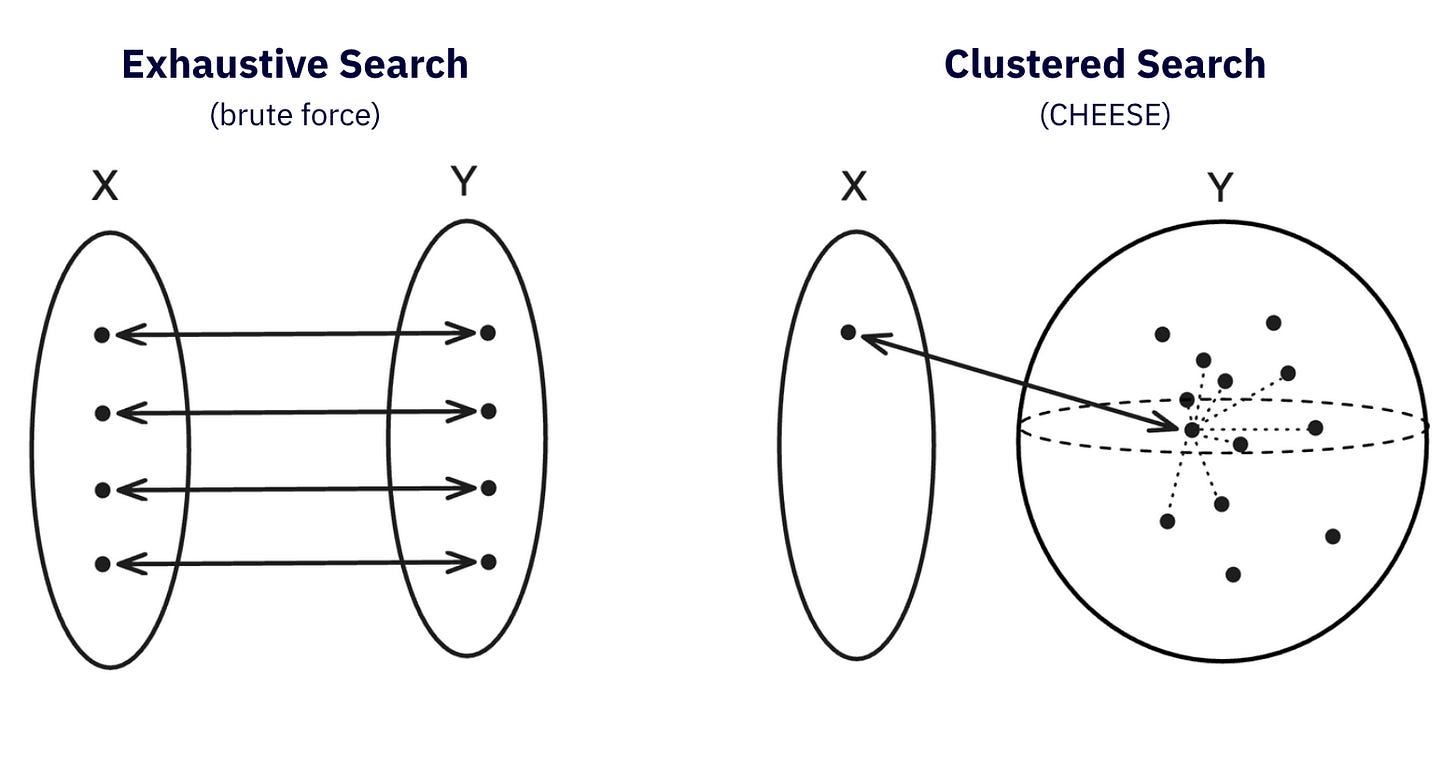
None of the existing solutions on the market is a silver bullet: docking is highly limited by the size of the database and resources: a no-go on billion scale databases. Fingerprint search (ECFP, Morgan, MACCS…) is computationally feasible, but cannot fully capture the shape or electrostatics of a molecule nor the binding site. Then there are 3D shape matching methods (ROCS, Quick Shape or eSim…): they work with the actual conformers and their volumetric overlaps, which makes it more physicochemically accurate, but also slow, or requiring huge GPU clusters and astronomic budgets to actually scale.
💡The Idea
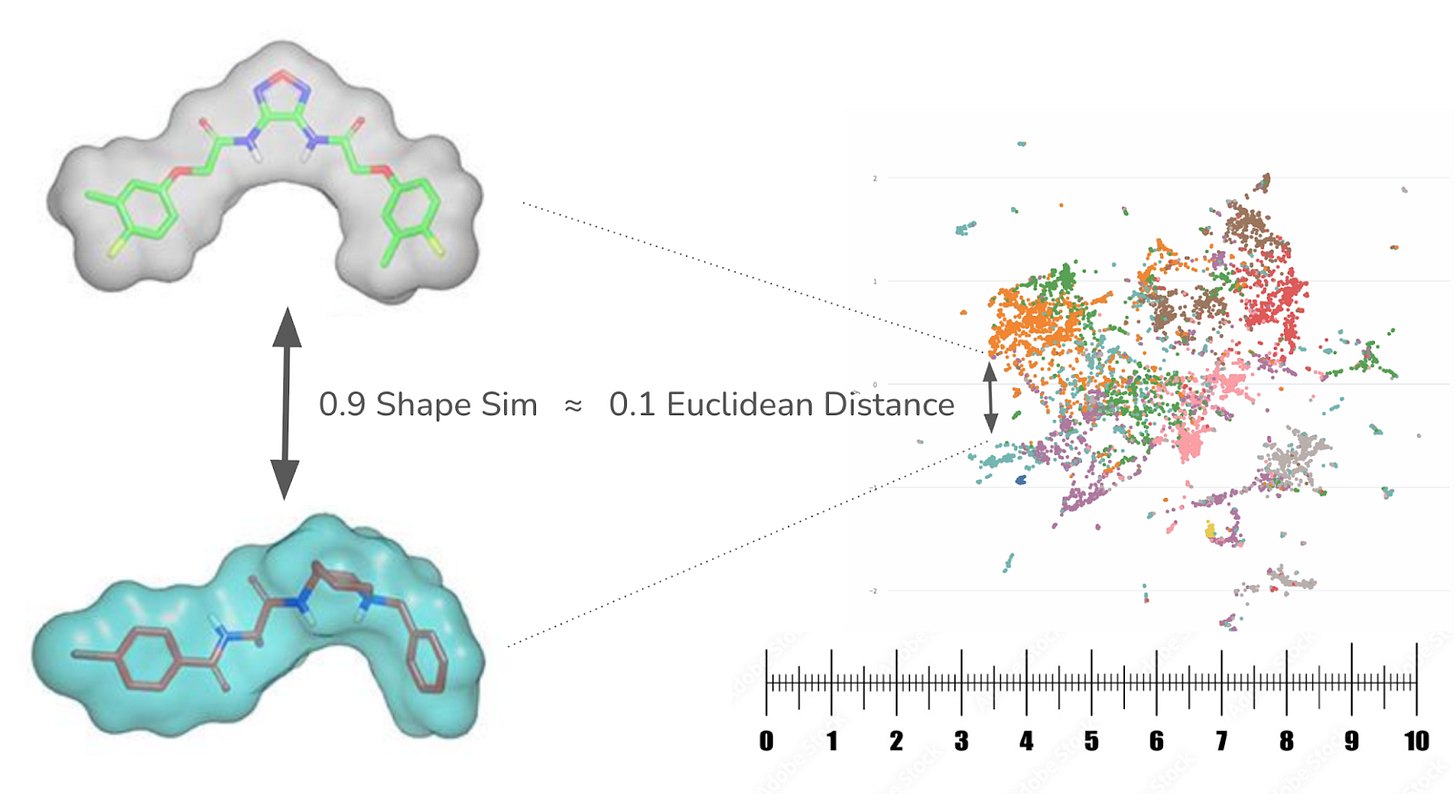
Deep MedChem asked a simple question: What if you could keep the precision of real 3D shape and electrostatics while searching at the speed of a modern vector database?
The result is CHEESE, the Chemical Embeddings Search Engine. Instead of running brute-force pairwise shape comparisons, CHEESE learns to embed molecules in a high-dimensional vector space where Euclidean distance represents true 3D shape and electrostatic similarity.
The team trained CHEESE on millions of molecule pairs from the ZINC15 database, aligning entire conformer ensembles and teaching the system to match their actual 3D overlap. Once deployed, it only needs a canonical SMILES input to place a molecule into this learned vector space. No extra conformer sampling or alignment is needed at search time.
This means CHEESE can screen billions of molecules for likely 3D matches in seconds on a standard CPU, increasing the scalability by three orders of magnitude compared to existing methods.
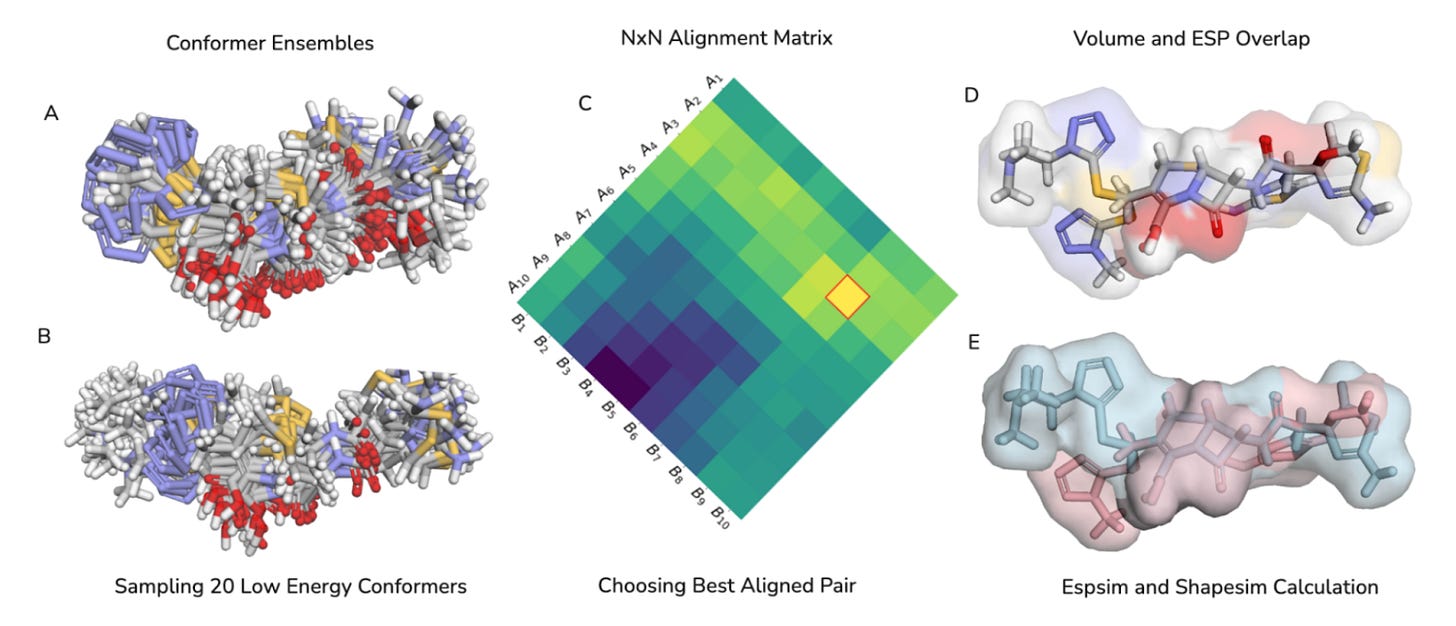
🔬 Why It’s Different
CHEESE stands out because it combines rigorous 3D physics with modern metric learning. Instead of relying on simple 2D fingerprints, it preserves detailed 3D shape and electrostatic features (Shapesim and Espsim) inside a learned vector space that works across real, diverse chemical libraries.
Benchmark tests show CHEESE matches or outperforms state-of-the-art 2D and 3D screening tools:
Up to 1000 times faster than traditional 3D shape matchers like ROCS.
Significant cost savings by screening the entire Enamine REAL database on a single 4-core CPU and SSD instead of a large GPU cluster.
Proven generalization across twelve different chemical spaces, from ZINC and ChEMBL to FooDB and COCONUT, with reliable results even for molecules it was not trained on.
Currently covering over 30 Billion molecules of enumerated commercial chemical space including, Enamine REAL, Mcule, eMolecules, Chemspace, Chemriya or MolPort databases in matter of seconds.
🔮The Future
DeepMedChem’s roadmap expands CHEESE into a full suite. Explorer, Modeller and Electrostatics are already available, giving researchers tools to visualize, generate and refine candidates within the same framework.
An open API also makes it possible to run large-scale similarity searches programmatically and integrate CHEESE directly into custom pipelines.
This vector approach could also support more advanced AI pipelines in the future, such as generative design workflows that benefit from instant 3D similarity feedback when exploring new molecules.
CHEESE: https://cheese-new.deepmedchem.com/
Deep MedChem: https://deepmedchem.com/
Paper: https://chemrxiv.org/engage/chemrxiv/article-details/67250915f9980725cfcd1f6f