🧬 GenomePAM: Letting the Human Genome Discover PAMs for You
Deep Dive | Edition 11
Welcome back to the deep dive, where we break down the AI tools and data reshaping how new drugs are discovered. In each edition, we speak directly with the teams behind these tools to explain what they solve, how they work and where they are going next.
What’s your biggest time sink in early drug discovery process?
Today we’re having a look at GenomePAM, a method from Zongli Zheng’s lab that addresses one of CRISPR’s most notoriously stubborn challenges: figuring out PAM requirements for different Cas enzymes. These short DNA sequences decide whether a Cas enzyme can recognise and bind to its target but discovering them in mammalian cells has long been a slow, messy and expensive process.
GenomePAM takes a different path. By turning naturally repetitive sequences across the entire genome into a built-in PAM library, it allows direct discovery in mammalian cells. So, no protein purification, no synthetic oligos, just an identical set of thousands of real targets per cell.
It’s a pretty innovative solution: just let the genome do the work. Although, as we found out, what appears to be a simple switch required a lot of hard work and hustle from Zheng and his team.
🔴 The Problem
Every new Cas enzyme comes with its own PAM code. Without knowing that code, you don’t know where the enzyme can actually bind in the genome, which is what makes PAM discovery the first and ultimately unavoidable step when exploring new nucleases.
The trouble is, the standard ways of doing this have long been painfully inefficient. In vitro assays demand purified protein, which can be hard or impossible to obtain for many novel or engineered Cas variants. Tube-based digestion methods require incredibly tight kinetic control, otherwise the enzyme can end up cutting indiscriminately. Even the “no purification” kits that express protein in a tube are seriously expensive and difficult to scale.
Cell-based screens aren’t much better at all. Most rely on inserting synthetic PAM libraries into cells but each cell only carries a tiny fraction of the library, or below one candidate per cell (multiplicity of infection [M.O.I] << 1). This means you need millions of cells and repeated rounds of screening to cover enough diversity and because expression levels vary from cell to cell, the results often require heavy normalization. Not ideal.
💡 The Idea
Zheng explained to us how his team had tried many of the existing approaches from protein purification to in-tube digestions, and even costly expression kits but each one proved too complex, too resource-intensive, or too unreliable to be practical long term.
So, Zongli Zheng and his team asked themselves a simple question: what if PAM discovery didn’t need synthetic libraries or protein purification at all? Could it be possible to use the human genome as the library instead?
The team targets a common 20-base repeat called Rep-1. It shows up 8,471 times in the reference human genome, which is about 16,942 copies per diploid cell. Each copy sits next to different surrounding bases. That variety gives you thousands of natural PAM contexts to test with one guide RNA.
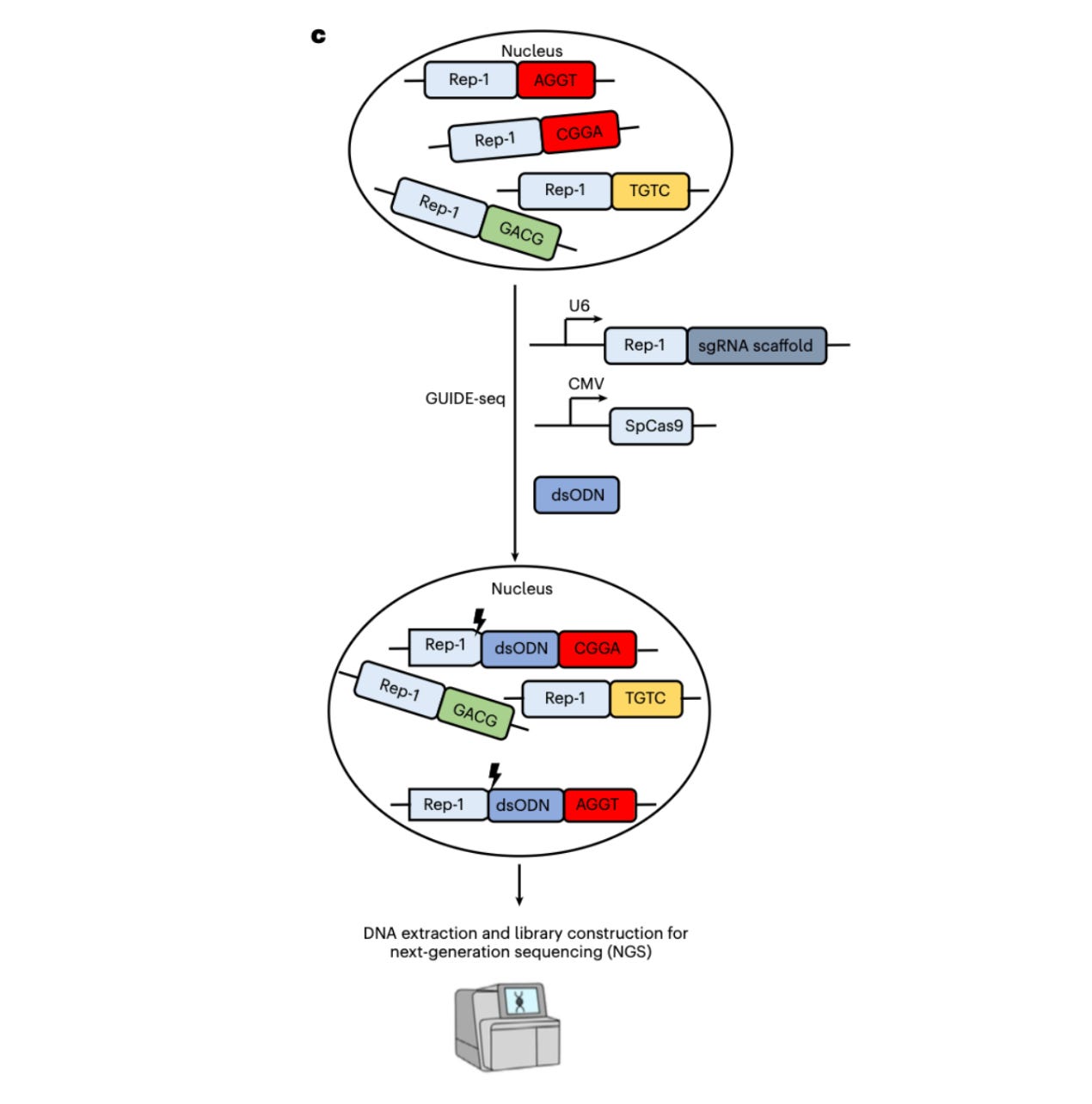
Here is what’s happening in the assay:
You express a Cas enzyme and a guide that points to Rep-1 (or to the reverse-complement for enzymes with 5′ PAMs).
Wherever a compatible PAM is present, the enzyme cuts.
Those cuts are “tagged” using GUIDE-seq, which inserts a tiny DNA tag at break sites.
Sequencing the tagged sites tells you which PAMs the enzyme actually used inside living human cells.
Because this can be run directly in mammalian cells, you avoid the big pain points: no protein purification, no tricky tube kinetics, and no huge synthetic oligo libraries. You get a direct readout of PAM requirements, plus a view of tolerated mismatches, all in the right chromatin context. Genius!
The turning point came when one of his students tested GenomePAM for the first time. The data was clean and very accurate, showing that PAMs could finally be characterized directly in human cells without all the extra layers of infrastructure. You can only imagine how exciting this must have been for the GenomePAM team.
🔬 Why It’s Different
GenomePAM matters because it moves PAM discovery into the place that counts most: living human cells. It trades months of tedious setup for a simple, scalable readout, turning a single guide into evidence across thousands of genomic contexts. The result is faster learning, cleaner comparisons, and a clearer path to pick the right nuclease for the job at hand.
Key changes scientists will notice about GenomePAM:
Directly in human cells. Reads PAM preferences in native chromatin; no protein purification, no synthetic oligo libraries.
Scales by design. Uses common genome repeats (for example, Rep-1) so one sgRNA probes thousands of contexts in a single assay.
Grounded and generalizable. Reproduces canonical PAMs (SpCas9, SaCas9, Cas12a) and handles hard cases like SpRY and CjCas9 in the same framework.
Built for comparison. Measures potency on thousands of matches and specificity across tens of thousands of mismatches, letting you rank nucleases and variants at scale.
Enables discovery and engineering. Supports finding active new nucleases and guiding PAM engineering within the same assay.
Large PAM window. While previous methods typically look for 4 to 6-base window PAM, GenomePAM easily checks for at least 10 bases in one go.
🔮The Future
What’s the future of GenomePAM and Zongli Zheng’s team? They want to tackle another enormous challenge in CRISPR-based therapies: off-target. To get there the team wants to compare PAM and off-target simultaneously to see which PAM offers the best trade-off of efficacy and off-targets.
📄 Read the paper!
⚙️ Access the model on Github.
👨🔬 Get in touch with Zongli.
Thanks for reading Kiin Bio Weekly!
💬 Get involved
We’re always looking to grow our community. If you’d like to get involved, contribute ideas or share something you’re building, fill out this form or reach out to me directly.
Subscribe now to stay at the forefront of AI in Life Science and keep up with this upcoming season of deep dives.
Connect With Us
Have questions on this or suggestions for our next deep dive? We’d love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website


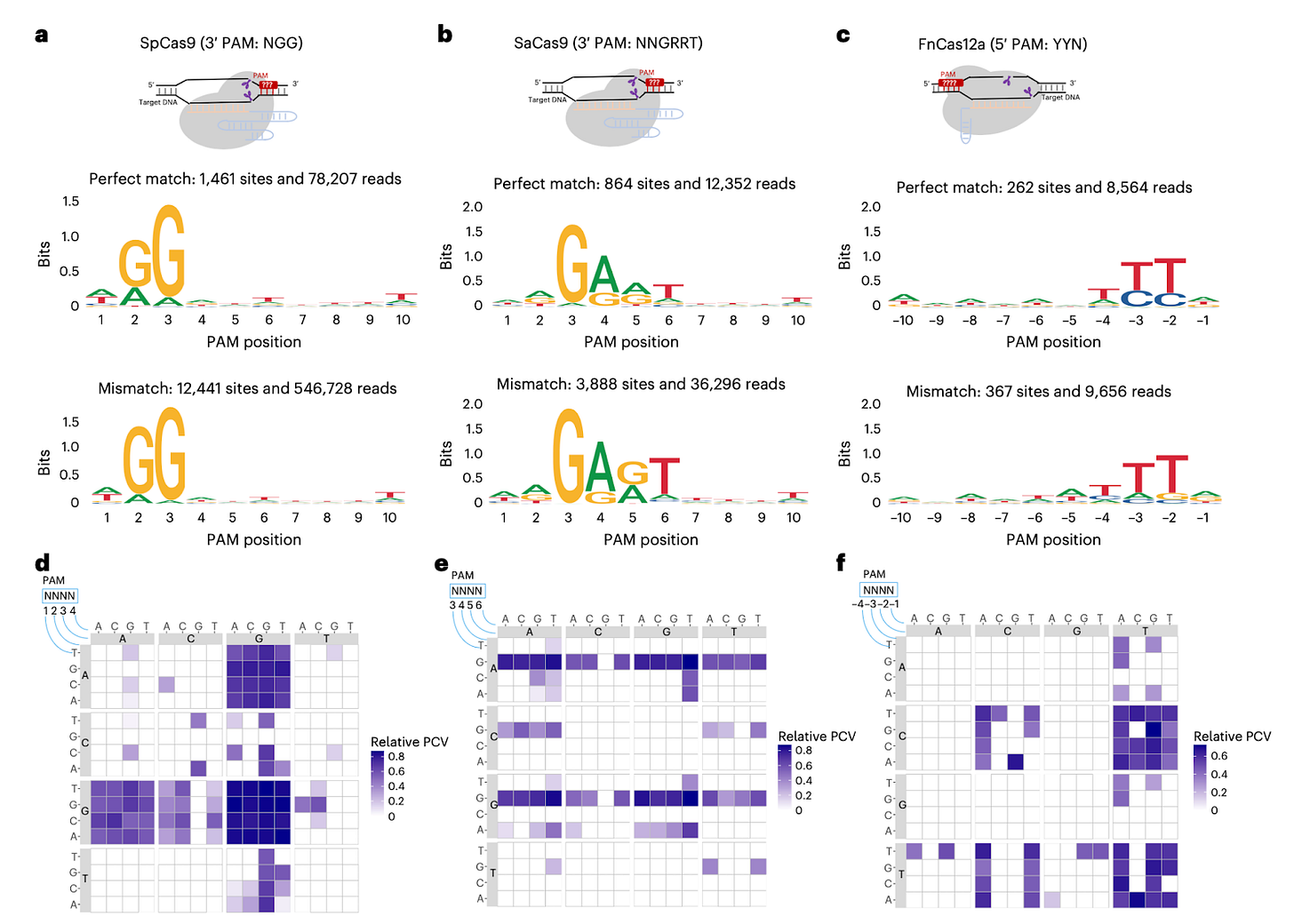
Impresssive methodological flip on PAM discovery. Using Rep-1's natural distribution acros the genome instead of synthetic libraries elegantly sidesteps the scalability and normalization problems that plague traditional screens. What stands out is how this shifts PAM characterization from a bottleneck into a parallelized readout, essentially letting chromatin context validate the enzyme in situ rather than extrapolating from tube conditions. Makes you wonder how many other discovery workflows are just waiting for a similar genome-as-resource inversion.