Germinal: AI for Epitope-Targeted Antibody Design
Deep Dive | Edition 9
Welcome back to the deep dive, where we break down the AI tools and data reshaping how new drugs are discovered. In each edition, we speak directly with the teams behind these tools to explain what they solve, how they work and where they are going next.
This week we’re looking at Germinal, a generative AI framework for antibody and nanobody design from Stanford University and the Arc Institute. Its lead developer, Luis Santiago Mille Fragoso, is a fifth-year PhD student in Bioengineering whose work sits at the intersection of structural biology and machine learning.
“After spending years designing biology in the wetlab, I realised how slow and unpredictable it could be,” Santiago said. “That’s what pushed me to look for ways to make it faster with computation”
Germinal is the result of that shift. It brings together structure prediction and language modelling to design new antibodies entirely in silico, targeting specific epitopes with just a handful of experimental tests.
🔴 The Problem
Antibodies are the workhorses of modern medicine, capable of binding targets with exquisite specificity. But discovering new ones is painfully slow. Traditional workflows rely on animal immunisation or massive library screens, testing millions of variants to find a few that bind, and almost never to a defined epitope.
Computational design promised a shortcut, yet progress lagged behind small-protein design. Antibodies are tricky: their complementarity-determining regions (CDRs) are flexible and disordered, confusing the structure predictors that powered recent breakthroughs. Most AI antibody generators either needed an existing weak binder or achieved only micromolar affinities after large-scale screening.
“There were great models for proteins in general, but nothing that could really imagine antibodies efficiently,” Santiago told us. “We wanted to fix that, to make something that could design them intelligently from scratch.”
💡 The Idea
The team’s insight was to combine two distinct kinds of intelligence: a structure predictor, AlphaFold-Multimer, and an antibody-specific language model, IgLM. Each offers a different skill. Structure models know what fits, while language models know what looks natural. The challenge was getting them to cooperate.
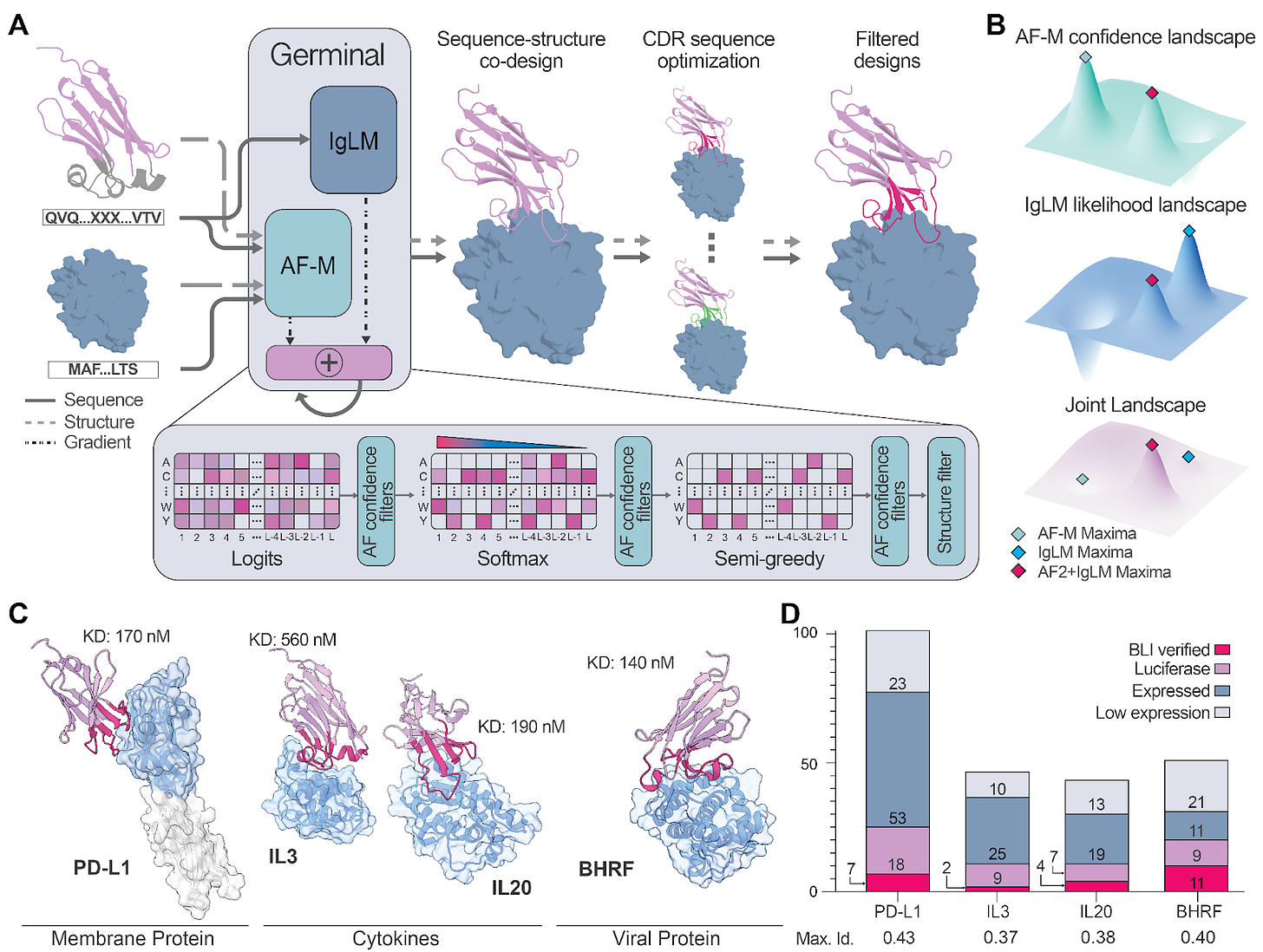
“At one point we almost gave up,” Santiago admitted. “We tried versions that only used AlphaFold without any modification and nothing worked. Then we saw a preprint showing that mixing a structure model with a language model improved antibody sequence recovery. That was the spark.”
Germinal achieves this through gradient merging, a method for blending the optimisation signals from both models. Gradients from AlphaFold-Multimer push toward sequences predicted to form stable, epitope-specific complexes, while those from IgLM bias the design toward real antibody patterns such as framework conservation, realistic CDR composition, and developability.
“Think of it like two experts arguing,” he explained. “AlphaFold tells you what looks physically stable; the language model tells you what looks biologically reasonable. Gradient merging is just teaching them to compromise.”
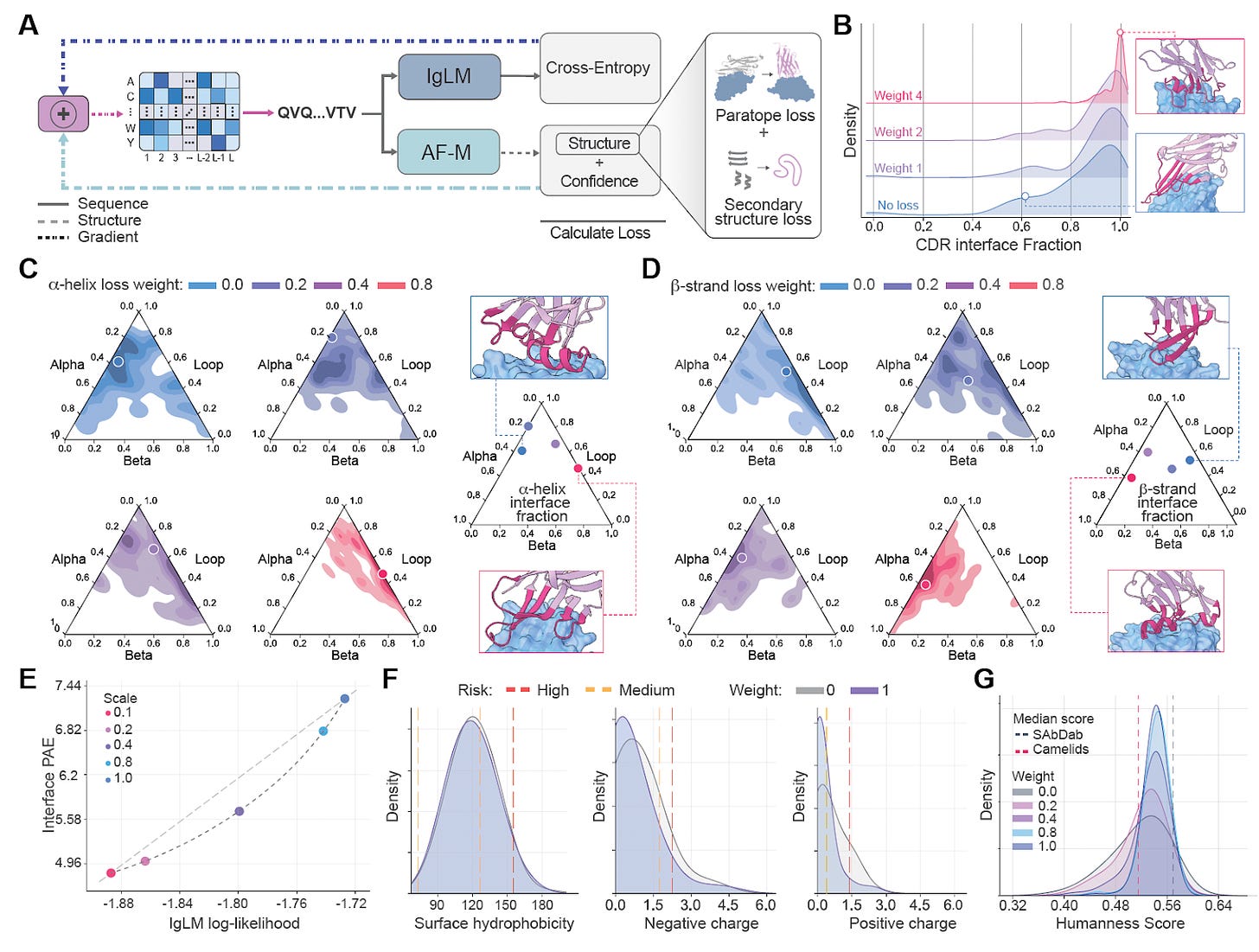
Balancing these forces revealed a Pareto frontier between structural confidence and antibody naturalness. By adjusting IgLM’s weight, the team could trade a bit of AlphaFold certainty for improved humanness and manufacturability, something not previously quantified in antibody design.
Germinal runs in three stages:
Design: joint optimisation of sequence and structure guided by merged gradients.
Sequence optimisation: refinement with AbMPNN, a structure-aware design model tuned for antibodies.
Filtering: re-evaluation with AlphaFold 3 and PyRosetta metrics to rank candidates.
Custom loss functions ensure binding occurs through CDRs rather than frameworks, and that loops remain flexible instead of collapsing into helices or beta strands.
The result is a generative system that starts from any target structure, defines the binding region, and proposes full nanobody or scFv sequences likely to hit that epitope, all without retraining.
📊 What About the Training Data
Unlike most AI models, Germinal does not train new networks. It reuses pre-trained weights from AlphaFold 2/Multimer for structure and IgLM, which was trained on large antibody repertoires such as OAS and SAbDab. The novelty lies in how these systems are used together.
Each design begins from an antibody framework, with CDRs free to vary under joint optimisation and framework biased toward the original sequence. Candidates are then filtered using AlphaFold 3 confidence metrics (pLDDT, ipTM, PAE) and PyRosetta features such as hydrogen bond counts, number of interface residues and interface shape complementarity.
Experimentally, the team validated designs against four antigens, PD-L1, IL-3, IL-20, and BHRF1, testing only 43-101 designs per target. Yet 4-22% bound successfully with nanomolar affinities (140-560 nM) and expressed well in mammalian cells.
“We got most of our results in about two months,” Santiago laughed. “It was an insane sprint. The day the first binder worked in the lab, everyone went nuts.”

Low-throughput success was enabled by a split-luciferase assay for rapid screening, followed by bio-layer interferometry (BLI) for kinetics. Every step, from code to protocols, is open source.
🔬 Why It’s Different
Germinal shifts antibody design from screening to reasoning.
Epitope-targeted generation: users can specify exactly where to bind.
Multi-objective optimisation: structure, sequence plausibility, and developability optimised in one loop.
Low-throughput success: nanomolar binders from under a hundred designs per target.
Interpretability: the Pareto frontier lets researchers steer designs.
“The first time we saw a real binding curve, after running everything in silico, it was wild,” Santiago said. “We went from pure code to a molecule that actually binds.”
🔮 The Future
The team is now expanding Germinal toward full-length antibody (scFv) design, with early results matching nanobody success rates. They are also extending the testing for epitope specificity, confirming that generated binders truly hit intended sites.
Longer term, Germinal could compress antibody discovery from months to days. Integrating it with diffusion-based backbone generators or reinforcement learning could make the process faster and more general.
“What excites me most isn’t just therapeutics,” Santiago said. “It’s the idea that any lab could design an antibody to study a protein the same week they discover it. That’s the dream.”
Germinal shows that epitope-targeted de novo antibody design is no longer science fiction. It is a reproducible, open-source reality emerging from the intersection of structure prediction, language modelling, and creative computational engineering.
📄 Read the paper!
⚙️ Access the model on Github.
Thanks for reading!
Did you find this newsletter insightful? Share it with a colleague!
Subscribe now to stay at the forefront of AI in Life Science and keep up with this upcoming season of deep dives.
Connect With Us
Have questions on this or suggestions for our next deep dive? We’d love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website

