🧫 IGLOO: Tokenising the Language of Antibodies
Deep Dive | Edition 10
Welcome back to the deep dive, where we break down the AI tools and data reshaping how new drugs are discovered. In each edition, we speak directly with the teams behind these tools to explain what they solve, how they work and where they are going next.
What’s your biggest time sink in early drug discovery process?
Today we’re diving into IGLOO, the ImmunoGlobulin LOOp Tokeniser, a new framework from Genentech’s Prescient Design team that rethinks how antibody loops are represented and modelled.
We spoke with Frédéric Dreyer, a researcher at Genentech and one of IGLOO’s creators, whose work bridges structural biology and machine learning. The project began as a three-month collaboration with Ada Fang from Maria Žitnik’s lab at Harvard, but its implications reach far beyond a summer project.
🔴 The Problem
Antibodies are nature’s most adaptable binding molecules. They owe that flexibility to their loops: short segments known as complementarity-determining regions (CDRs) that form the business end of every antibody.
Despite decades of study, modelling these loops remains one of the toughest open problems in antibody engineering.
For over 30 years, researchers have used canonical clusters to group CDRs by backbone geometry. It worked for some loops but broke down for others. Nearly three-quarters of H3 loops remain unclassified, and these clusters ignore sequence entirely, making them incompatible with modern protein language models (PLMs) that learn only from amino acid sequences.
“There’s this gap,” Dreyer said. “Structure-based methods know what antibodies look like, and language models know what they say. But there was no shared vocabulary between the two.”
That disconnect makes it difficult to build models that leverage all of the available data while reasoning over both sequence and structure, the essential link for antibody design and optimisation.

💡 The Idea
IGLOO addresses this bottleneck by tokenising full antibody loops rather than individual residues, treating each loop as a meaningful structural phrase in the antibody language.
“Most structure models represent proteins residue by residue,” Dreyer explained. “IGLOO encodes entire loops, segments that are biologically meaningful because they directly contact the antigen.”
Instead of assigning each residue a structural token, IGLOO represents the full loop as a single multimodal embedding, combining sequence and backbone geometry. Loops with similar 3D shapes and sequence patterns sit close together in the model’s embedding space, while dissimilar ones are pushed apart.
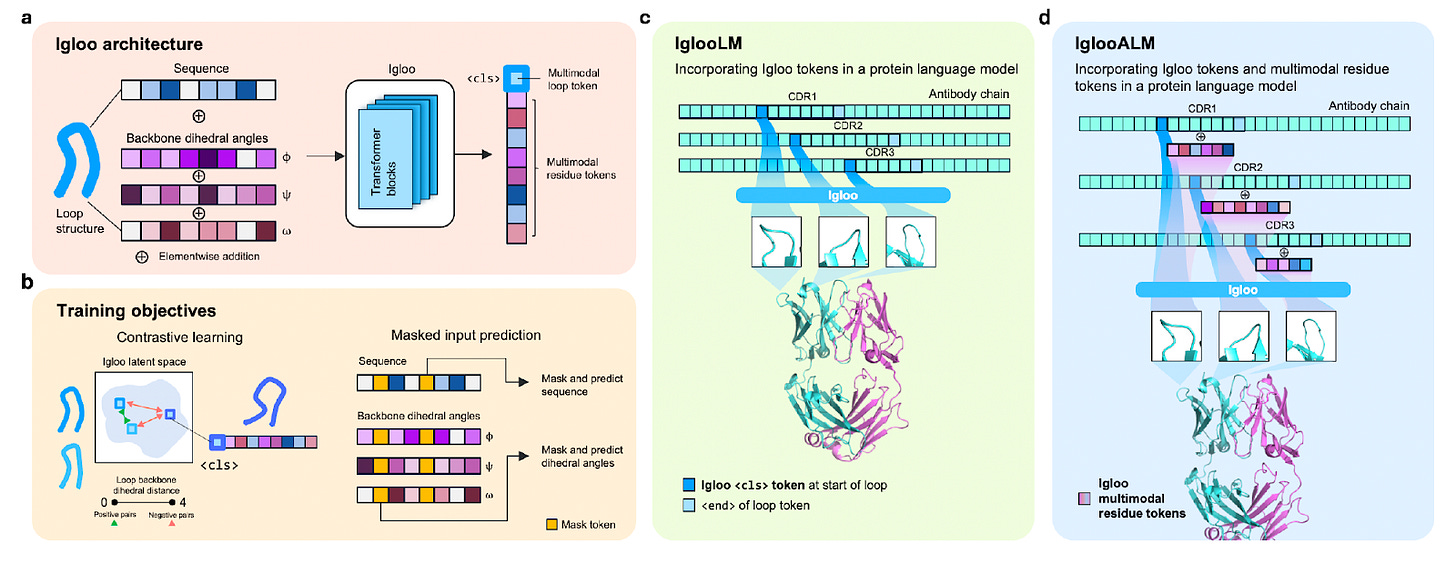
The key is a contrastive learning objective built on dihedral angles rather than RMSD. By comparing loops based on their angular geometry, IGLOO learns a geometry-aware “loop language” that captures subtle structural variation.
The result is that every loop, canonical or novel, gets a token that reflects both its shape and sequence, creating a shared language that works seamlessly with antibody LMs while staying physically grounded.
📊 The Data
To build that shared space, IGLOO was trained on around 15,000 experimental antibody and T-cell receptor structures from SAbDab and STCRDab, augmented with approximately 100,000 predicted structures generated from paired sequences in the Observed Antibody Space (OAS).
Each loop was represented by its amino acid sequence and backbone dihedral angles (ϕ, ψ, ω). The model learned to reconstruct masked residues and angles, align structurally similar loops, and discretise its embeddings into a codebook of loop tokens for retrieval and downstream use.
Once trained, the team integrated these tokens into antibody language models, producing IGLOOLM (loop-level tokens) and IGLOOALM (loop and residue tokens). Both were fine-tuned from IgBert, an antibody LM pre-trained on roughly two billion sequences, then refined on a mix of experimental and predicted structures.
🔬 Why It’s Different
IGLOO represents a shift in how antibodies can be represented, searched, and designed.
1️⃣ Complete Coverage: Unlike canonical clusters, IGLOO can tokenise every loop, including unseen or atypical ones. It reproduces known canonical forms with high accuracy while expanding coverage across the antibody landscape.
2️⃣ Structural Retrieval: In benchmark tests, IGLOO achieved state-of-the-art precision for loop-structure retrieval, outperforming existing encoders. It improved accuracy on heavy-chain H3 loops by 5.9% compared with the best prior model.
3️⃣ Better Downstream Predictions: When embedded into antibody language models, IGLOO tokens improved performance. On AbBiBench, IGLOOLM outperformed its base model on eight of ten antibody-antigen pairs, matching models several times larger.
4️⃣ Controlled Loop Design: Using IGLOOALM, the team could generate new loop sequences that preserved their structural fold. In one example, redesigned SARS-CoV-2 H3 loops achieved <1 Å RMSD with only 27% sequence identity, showing strong structural fidelity and generative control.
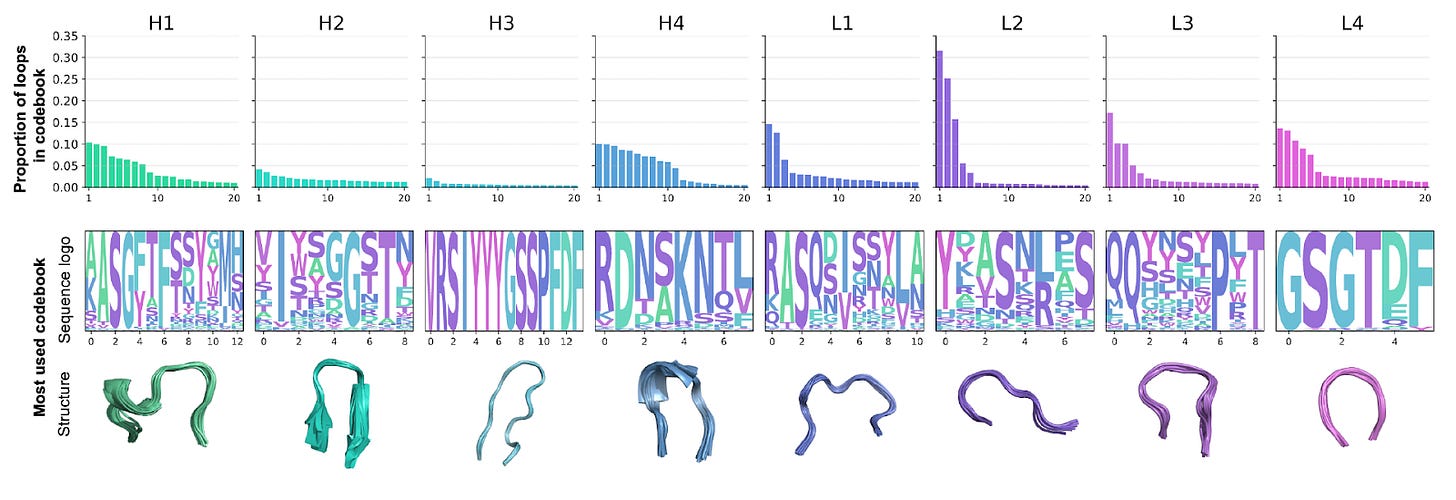
“The embedding space was trained unsupervised,” Dreyer said, “yet it rediscovered the canonical clusters structural biologists had spent decades mapping. It was like watching the model learn antibody biology from scratch.”
🔮 The Future
For Dreyer, IGLOO is more than a single model. It is a blueprint for the next generation of antibody foundation models.
“What excites me is bringing structure and sequence into a common language,” he said. “We have billions of antibody sequences but only tens of thousands of structures. IGLOO shows we can connect those worlds in a scalable way.”
The next frontier is multimodality: models that integrate sequence, structure, dynamics, and antigen context into a unified framework. Within Genentech, similar representations are already being tested for binder optimisation, developability prediction, and affinity maturation.
Dreyer sees IGLOO-style embeddings becoming the backbone of generalist antibody-design systems, models that understand not just what sequences look like, but how they behave.
“I think of it as merging the hard-won domain knowledge from antibody biology with the scalability of modern machine learning,” he said. “That is how we get to truly generalisable design systems.”
📄 Read the paper!
⚙️ Access the model on Github.
👨🔬 Get in touch with Frederic.
Thanks for reading Kiin Bio Weekly!
💬 Get involved
We’re always looking to grow our community. If you’d like to get involved, contribute ideas or share something you’re building, fill out this form or reach out to me directly.
Subscribe now to stay at the forefront of AI in Life Science and keep up with this upcoming season of deep dives.
Connect With Us
Have questions on this or suggestions for our next deep dive? We’d love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website

