Manifold Bio’s mBER, UF’s GatorAffinity, and Google’s PHA
Kiin Bio's Weekly Insights
Welcome back to your weekly dose of AI news for Life Science!
What’s your biggest time sink in the drug discovery process?
mBER: Controllable de novo antibody design platform with million-scale experimental screening
Antibody design usually means compromise. You can generate unconstrained minibinders by the thousands, but if you want something usable, like a nanobody with therapeutic potential, your design space shrinks fast.
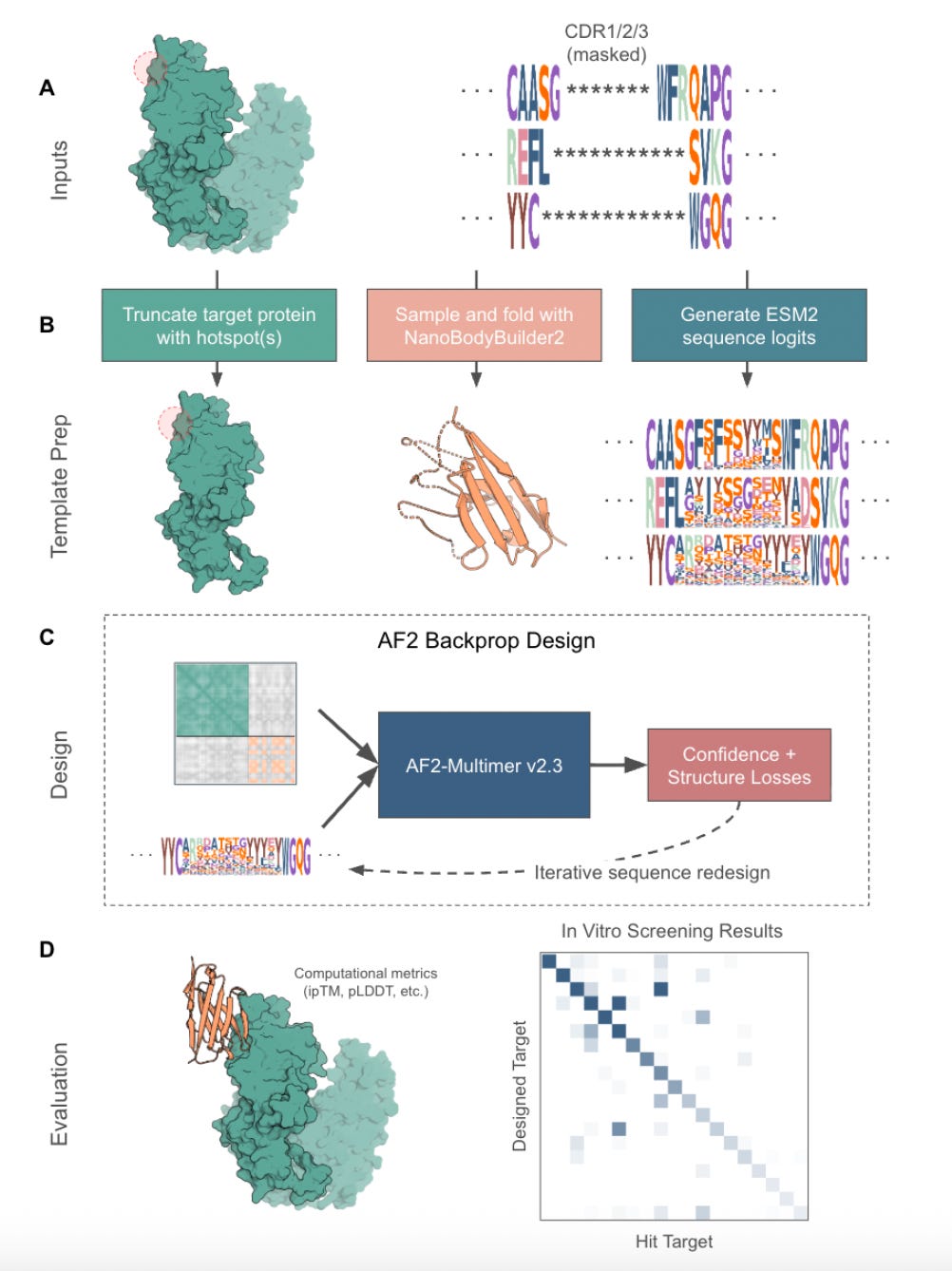
That’s what makes mBER so interesting. Built by Manifold Bio, mBER is a smart, format-aware wrapper around existing models like AlphaFold-Multimer and ESM2. It generates VHH binders that respect the structure and sequence constraints of real nanobodies, without any model fine-tuning or custom pretraining. Just smart templating, efficient search, and a lot of screening.
The proof is in the experimental pudding. The team designed over one million VHH sequences across 436 targets and screened them against 145 antigens, generating a dataset of more than 100 million protein-protein interactions. Sixty-five targets showed significant hits, and some epitopes reached hit rates of 38%.
Applications and Insights
1. Format-aware binder generation
mBER guides AlphaFold-Multimer to design within a realistic nanobody scaffold. It uses fixed framework residues, masked CDRs, and ESM2 priors to generate structured, sequence-valid outputs that are usable in downstream pipelines.
2. Big-screen validation
More than one million designs were screened across 145 targets using phage display. Forty-five percent of targets yielded statistically significant hits, with several epitopes hitting success rates above 30 percent.
3. Filtering that works
AlphaFold’s ipTM score was a strong predictor of success. Filtering for ipTM > 0.8 improved hit rates up to 10 times, making it easier to prioritise sequences for synthesis.
4. Multi-epitope targeting
Several antigens yielded binders at multiple distinct epitopes, showing that mBER can explore different binding modes and generate functionally diverse hits for the same target.
What about the training data?
mBER doesn’t require model pretraining. Instead, it builds design context on the fly using existing tools:
Structure and sequence priors come from ESM2 and NanoBodyBuilder2, which provide scaffold templates and realistic amino acid distributions. CDRs are masked and sampled, while framework residues are fixed.
AlphaFold-Multimer is used as-is, without fine-tuning. Backpropagation through the model helps guide sequence selection toward desirable structures.
Experimental data comes from two phage display screens, covering more than one million sequences and over 100 million antigen-binder interactions.
All software and data are open-source and available to the community.
I thought this was cool because it’s a great example of what smart engineering can do with the tools we already have available to us. mBER doesn’t introduce a new model. It builds the scaffolding to make foundation models like AlphaFold and ESM2 useful in real therapeutic design contexts.
That might sound simple, but it is not. Designing antibody-like binders that actually fold, express, and hit diverse epitopes usually takes months. mBER shows that you can do it in days, using open models and accessible infrastructure. Now, with more than 100 million datapoints already generated, it sets the stage for a new kind of binder benchmarking, one where format, epitope, and affinity can all be explored systematically.
For anyone working on therapeutic design, protein engineering, or functional screening at scale, mBER is a toolkit worth watching for sure.
📄Check out the paper!
⚙️Try it out the code.
GatorAffinity: Large-scale synthetic structural pretraining for accurate protein-ligand affinity prediction
Most structure-based affinity prediction tools hit a wall: the PDBbind dataset, with fewer than 20,000 high-quality experimental complexes. That’s not much, especially when machine learning thrives on scale.
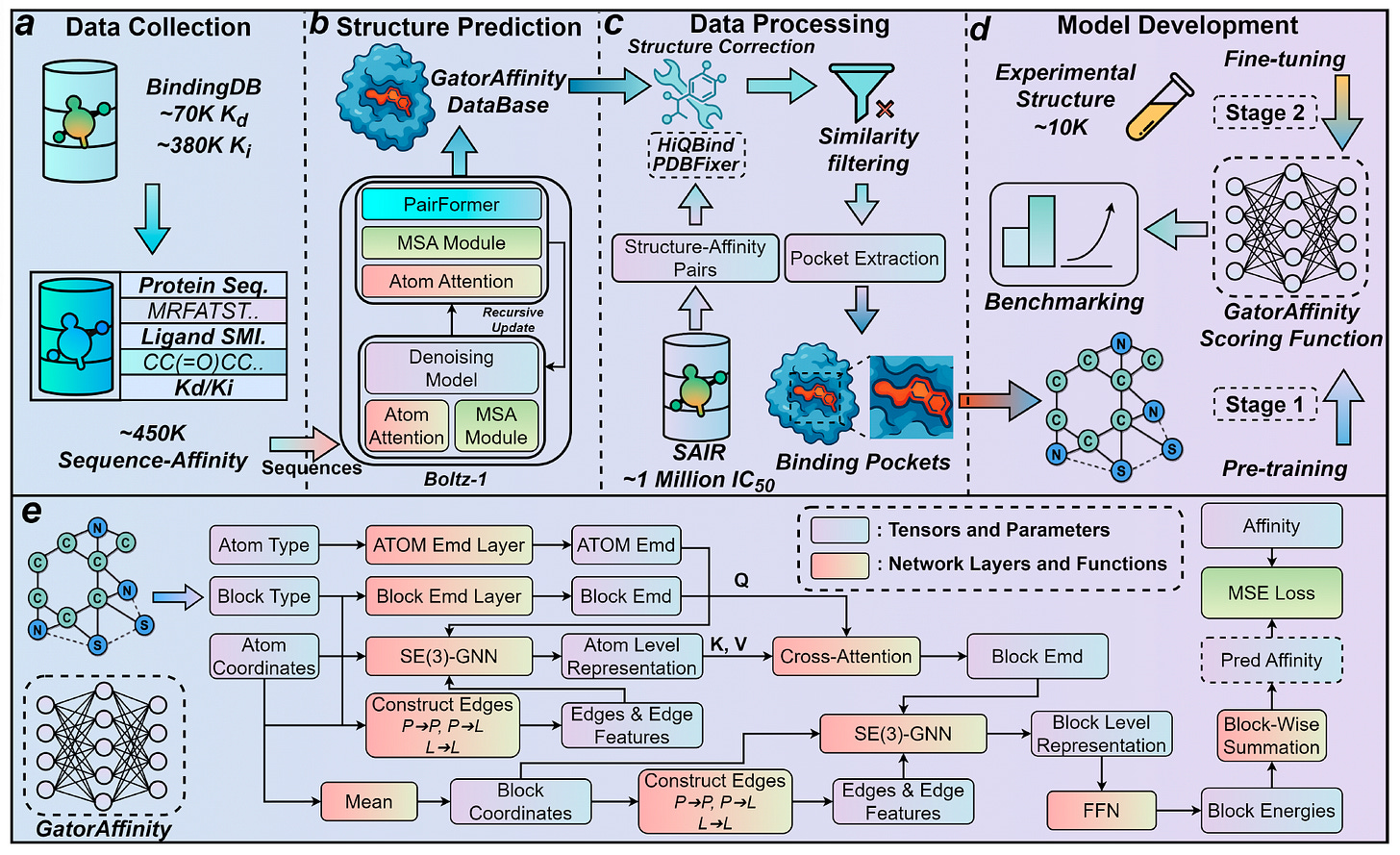
GatorAffinity breaks through that wall. Developed at the University of Florida, this new model is trained on more than 1.5 million synthetic protein–ligand complexes with annotated binding affinities, created using Boltz-1 and SAIR. With that kind of pretraining, and careful fine-tuning on experimental data, it outperforms every other method on the field’s toughest benchmarks.
It also introduces a key idea: synthetic structure–affinity data can unlock scaling laws in this domain, just like we’ve seen in language and vision. The more data you give it, the better it gets.
Applications and Insights
1. Breaks records on affinity benchmarks
On the filtered LP-PDBbind dataset, GatorAffinity achieves an RMSE of 1.29, a Pearson R of 0.67, and a Spearman ρ of 0.65. That’s a significant improvement over the previous best, GIGN, across all three metrics.
2. Scales with data, not just parameters
Performance kept improving as the dataset grew. The best results came from training on a large mix of Kd, Ki, and IC50-labeled entries, showing the value of both scale and biochemical diversity.
3. Robust to noisy structures
Even when including low-confidence structures without ipTM filtering, the model performed well. That makes it easier to build large datasets without aggressive pruning.
4. Pretraining and fine-tuning are both essential
Joint training on synthetic and experimental data didn’t work as well. The best approach was to pretrain on synthetic structures, then fine-tune on high-quality experimental data.
What about the training data?
GatorAffinity is built on one of the largest synthetic structure–affinity datasets to date.
Source and scale: 450,000 Boltz-1 predicted protein–ligand complexes were generated from BindingDB entries with Kd and Ki labels. Another 1 million came from SAIR using IC50 data.
Structure post-processing: Complexes were cleaned using HiQBind and PDBFixer to correct bond orders, protonation, and clashes. Drug-like ligands and structurally plausible pairs were retained.
Training strategy: The model was first pretrained on all synthetic complexes, then fine-tuned on around 10,000 high-quality experimental structures from PDBbind2020.
All datasets and code are open access.
I thought this was cool because it proves you can get state-of-the-art results without needing a bigger model. You just need better data. GatorAffinity turns synthetic structure-affinity pairs into a real asset, enabling large-scale learning in a field where data has always been the bottleneck.
This feels like a turning point. If we can trust and scale synthetic structural data like this, the path is open for faster screening, more accurate scoring, and smarter generative workflows across the board.
📄Check out the paper!
⚙️Try out the code.
PHA: Multi-agent health reasoning framework integrating data science, physiology, and behavioural coaching
Most health apps collect data. Some even tell you things about it. Very few actually understand you.
That is what Google’s research project prototype, The Personal Health Agent (PHA), is attempting to change.
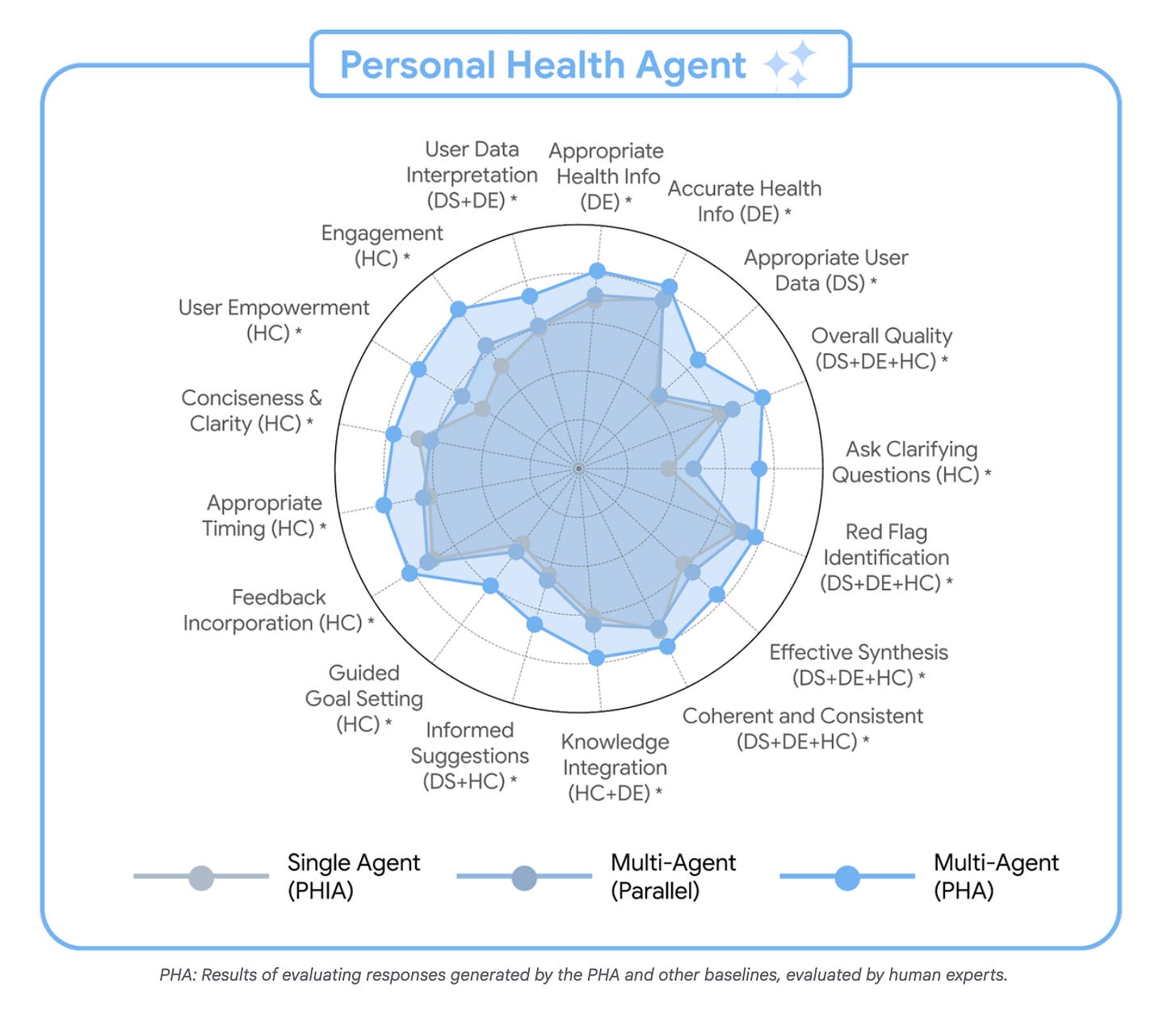
Instead of one model doing everything, PHA uses a multi-agent system of three AI specialists working like a care team:
A Data Science Agent that reads your numbers and finds patterns.
A Domain Expert Agent that explains why they matter.
A Health Coach Agent that turns these insights into action.
If you ask, “How did my workouts affect my sleep last month?” PHA does not just summarise your Fitbit graph. It finds correlations, explains the physiology, and gives you a plan that makes sense.
Not a chatbot. Not a wellness app. A digital scientist that reasons. Definitely will be interesting to see how this multi-agent team actually operates.
Applications and Insights
1. Built on real questions
The team analysed over 1,300 real user queries from Fitbit and Google Search to ground the system in what people actually ask about their health.
2. Collaborative reasoning
The agents share data and context dynamically, almost like a small research team combining analytics, biology, and behavioural science.
3. Extensive evaluation
Over 7,000 human annotations and 1,100 hours of expert review were used to assess PHA’s reasoning, interpretation, and coaching quality.
4. Grounded in real-world data
Tested on the WEAR-ME dataset, combining Fitbit metrics, lab results, and surveys from more than 1,100 participants.
What About the Training Data?
Importantly, no new user data was used to train models.
PHA runs on Gemini 2 within strict research boundaries, using only anonymised evaluation data.
That means your Fitbit data is not secretly training Google’s next health model.
It is a reminder that responsible AI design and scientific depth can coexist.
So it looks like PHA is not trying to be your doctor or your therapist. It’s aiming towards something in between, a system that sees your body as both data and biology. I think this is cool because it reframes what AI in health could be. Instead of telling you what to do, it helps you understand why your body behaves the way it does. It reasons, it explains, and it connects the dots. To me, that feels closer to how scientists think: health as a dynamic system that needs interpretation, not just fragmented tracking.
If projects like this keep evolving responsibly, AI might soon help people understand their own biology the way language models help us understand information: through reasoning, conversation, and context.
📄Check out the paper!
Thanks for reading!
Did you find this newsletter insightful? Share it with a colleague!
Subscribe Now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website