Microsoft’s Adaptive Resampling, IIT Madras’s PURE, and Bose Centre’s PathGennie
Kiin Bio's Weekly Insights
Welcome back to your weekly dose of AI news for life science. A big thank you for helping us reach 750 subscribers! It’s brilliant to see our community grow every week and have more people interested in the tools and software that we choose to spotlight.
What’s your biggest time sink in the drug discovery process?
🧬 Adaptive Resampling: Teaching AI to Learn from Rare Cells
What if single-cell models finally paid attention to the rarest and most biologically important cells?
Adaptive Resampling (AR), developed by Microsoft Research and collaborators, is a new training algorithm that tackles one of the biggest weaknesses in single-cell machine learning: models trained on heterogeneous datasets tend to ignore rare cell types even though these often drive disease progression and key biological processes.
Current approaches, including large foundation models, show 15 to 30 per cent performance drops on disease samples and miss around 20 per cent of rare populations. Manual annotation performs better but shows 25 per cent inter-annotator variability and does not scale.
AR reshapes training batches based on latent-space density. At each epoch, cells are projected into latent space and reweighted so that low-density (rare) cells are sampled more often, increasing their influence without increasing computational cost.
🔬 Applications and Insights
1️⃣ Improved Gene Expression Reconstruction
Across scVI experiments spanning six orders of magnitude imbalance, AR improved reconstruction accuracy using as few as 10 atlas cells. Standard training needed 1,000 cells to achieve similar results.
2️⃣ Superior Cell Type Classification
AR significantly outperformed standard training on heart, kidney and neuron datasets across F1, accuracy, recall and precision. Improvements appeared with only 100 training cells rather than the usual 10,000.
3️⃣ Generalisable Perturbation Prediction
On immune stimulation datasets across species, AR nearly doubled R² for several held-out groups, reduced MSE across the board, and produced cleaner latent embeddings that better preserved biological structure.
4️⃣ Model Agnostic and Metadata Free
AR works with any encoder-based neural network and requires no thresholds, metadata or architectural changes, making it widely applicable across single-cell tasks.
💡 Why It’s Cool
AR provides a practical way to teach models to learn from rare cells, improving performance in disease settings, small-sample regimes and out-of-distribution conditions.
It enhances generalisation while keeping training efficient through simple latent-space operations.
📄 Read the paper
💻 Try the code.
⚛️ PURE: A Synthesis-Aware Shift in AI Molecular Design
What if AI-generated molecules were not only interesting on paper but genuinely synthesizable in the lab?
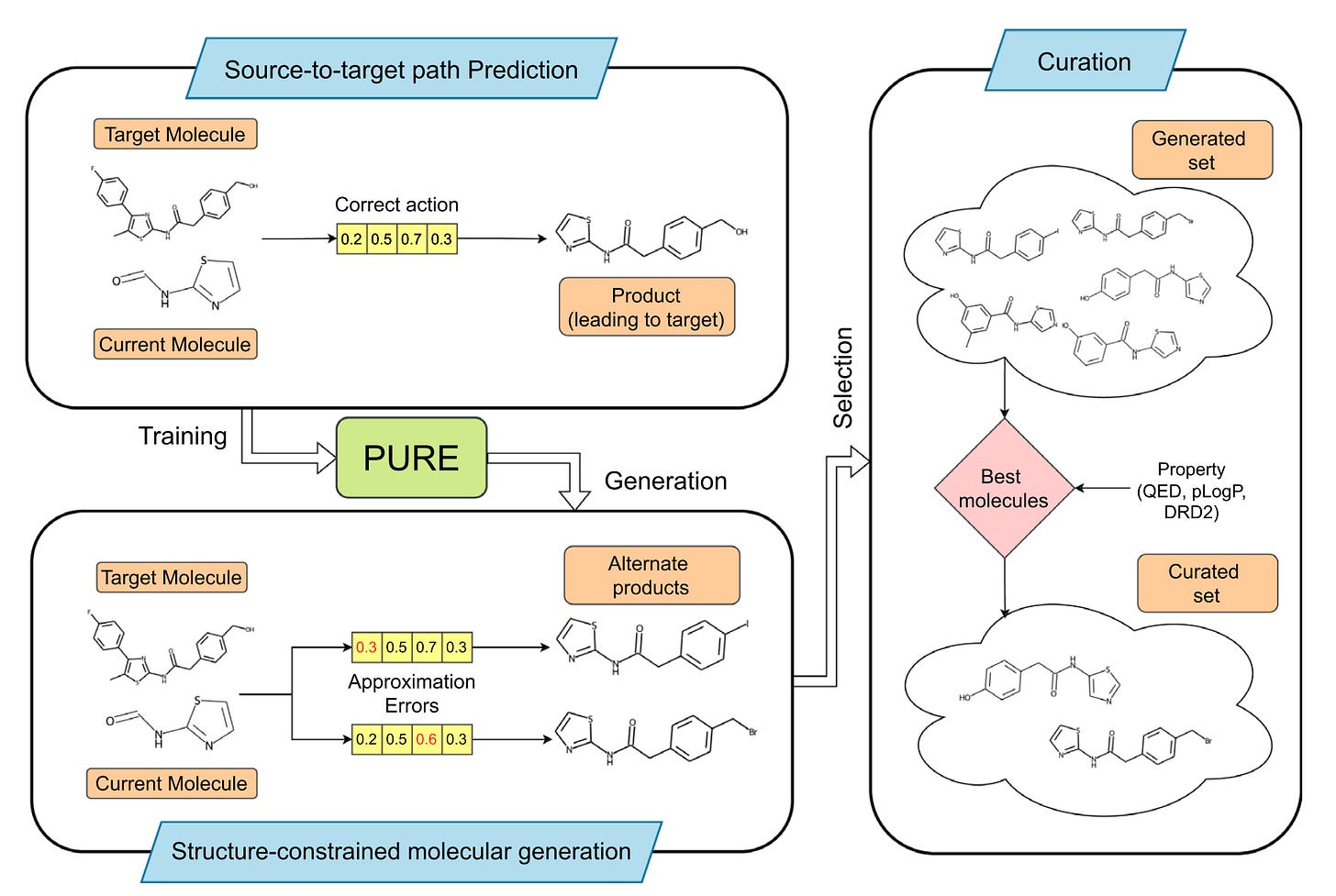
PURE, developed by researchers at the Indian Institute of Technology Madras and The Ohio State University, is a synthesis-aware generative model designed to answer the question most models ignore: could a chemist actually make this?
PURE, short for Policy-guided Unbiased Representations, generates molecules that reflect real chemistry and real reaction pathways rather than abstract numerical optimisation.
🔬 Applications and Insights
1️⃣ Molecule Generation That Mirrors Chemical Thinking
PURE uses policy-guided reinforcement learning to frame molecular design as a sequence of chemical actions. This is closer to how chemists plan synthesis than how traditional models optimise scores.
2️⃣ Built on Reaction Rules, Not Wishful Thinking
The system incorporates actual reaction logic. Every proposed structure maps to a plausible synthetic route. No imaginary chemistry.
3️⃣ Broad Exploration of Chemical Space
Unlike many models that collapse into small regions of chemical space, PURE explores widely and uncovers novel scaffolds.
4️⃣ Synthesis-Aware Suggestions
Every molecule is generated with an implicit sense of how it could be made, reducing the gap between virtual hits and real experiments.
⚡ Performance Highlights
Benchmarking on QED, DRD2 and solubility tasks showed that PURE consistently generated drug-like, synthesisable molecules instead of theoretical artefacts.
💡 Why It’s Cool
PURE represents a shift towards AI systems that understand chemistry, not just numerical optimisers.
It accelerates the path from computational design to real-world testing and grounds early discovery in practical medicinal chemistry.
📄 Read the paper
⚙️ Explore the code on GitHub.
Big thanks to our newest contributor, Yassir, for writing this week’s article on PURE. Great find! We’re looking forward to sharing more of your insights with our readers soon.
If you’re interested in contributing or joining the community, reach out to me or fill this out!
🧪 PathGennie: Fast, Unbiased Discovery of Rare Molecular Events
What if protein folding, ligand unbinding and other rare molecular events could be captured in tens of picoseconds?
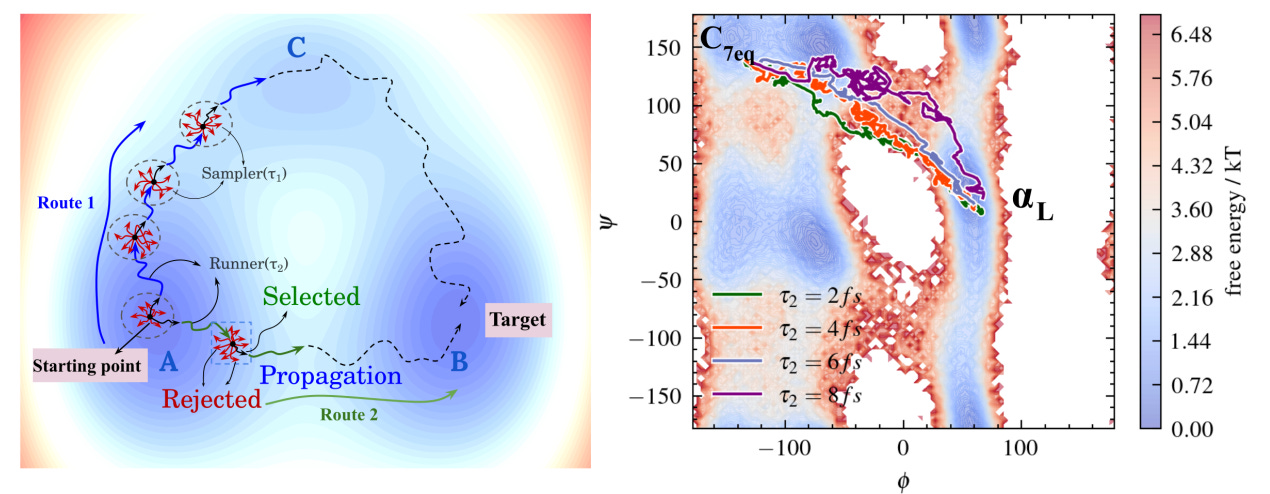
PathGennie, from the S. N. Bose National Centre for Basic Sciences, introduces an adaptive sampling framework that discovers transition pathways without biasing forces, elevated temperatures or long simulations.
The system launches ultrashort, unbiased molecular dynamics trajectories with randomised velocities and selectively propagates only those that make genuine progress in collective variable (CV) space.
The result is physically meaningful transition pathways generated extremely quickly while preserving the underlying dynamics.
🔬 Applications and Insights
1️⃣ Ligand Unbinding at Scale
PathGennie identified seven distinct benzene exit pathways in T4 Lysozyme L99A and recovered all three known imatinib escape routes from Abl kinase. It also uncovered low-frequency channels that biased methods often miss.
2️⃣ Fast Folding and Unfolding
For Trp-cage and Protein G, folding and unfolding events were generated in 150 to 600 ps and closely matched minimum free-energy paths obtained from long Anton simulations.
3️⃣ Robust to Suboptimal CVs
Even when provided with an intentionally poor one-dimensional CV on the Wolfe-Quapp surface, PathGennie still recovered multiple mechanistic routes, demonstrating strong resilience to CV choice.
4️⃣ Boosts Weighted Ensemble Performance
Seeding Weighted Ensemble workflows with PathGennie paths dramatically accelerated convergence:
• Alanine dipeptide: 1.7 microseconds reduced to 612 nanoseconds
• Chignolin: 1.4 microseconds reduced to 350 nanoseconds
💡 Why It’s Cool
PathGennie provides a simple, massively parallel and physics-faithful way to explore rare events.
It uncovers diverse mechanistic pathways, works with imperfect CVs and integrates seamlessly with WE, TPS and MSM workflows.
📄 Read the paper
⚙️ Code available on GitHub
Thanks for reading Kiin Bio Weekly!
💬 Get involved
We’re always looking to grow our community. If you’d like to get involved, contribute ideas or share something you’re building, fill out this form or reach out to me directly.
Connect With Us
Have any questions or suggestions for a post? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website