MSD’s Ovo, Helmholtz Munich’s scConcept, and Graz University’s Riff-Diff
Kiin Bio's Weekly Insights
Welcome back to your weekly dose of AI news for life science.
What’s your biggest time sink in the drug discovery process?
🧬 Ovo: Next-Generation Protein Design Made Accessible
What if de novo protein design were as simple as running a single command?
Modern protein design relies on fragmented pipelines that mix RFdiffusion, ProteinMPNN, AlphaFold, Rosetta and QC tools, often stitched together with custom scripts, HPC clusters and coding expertise. For many biologists, this makes state-of-the-art design inaccessible.
Ovo, developed by MSD Czech Republic, unifies the entire workflow into one open-source ecosystem. Backbone generation, sequence design, structure prediction and quality control all run through a single interface. Everything operates locally or on HPC without heavy setup or tool switching.
Ovo supports binder design, scaffold generation and protein ligand or protein protein interface modelling. Workflows are orchestrated through Nextflow and quality control is handled by ProteinQC, which computes more than 100 structural and sequence descriptors to filter designs early.
🔬 Applications and Insights
1️⃣ End-to-end design that scales
Benchmark campaigns generated realistic design sets:
4,800 oxidoreductase scaffolds leading to 13 accepted
5,000 PD 1 scaffolds leading to 275 accepted
4,000 insulin receptor binders leading to 5 accepted
All fully reproducible and managed through a single system.
2️⃣ High-throughput binder pipelines
Ovo integrates RFdiffusion with BindCraft. BindCraft produced 13 accepted binders in 6 hours on a single A10G GPU, making high-quality design possible on modest hardware.
3️⃣ Diversification improves hit rates
Partial diffusion refinement increased binder success from 0.075 per cent to 8.2 per cent, more than a 100 fold improvement without additional modelling complexity.
4️⃣ Built-in quality control
Across 361 accepted designs, ProteinQC metrics such as solubility, compactness and asphericity closely matched natural PDB proteins, indicating strong foldability and expression potential.
💡 Why It Is Cool
Ovo turns de novo protein design from a technical challenge into a practical tool. By lowering barriers and embedding quality control throughout the workflow, it makes modern protein design accessible to far more laboratories.
📄 Read the paper
⚙️ Explore the code on GitHub
🔬 scConcept: Teaching AI to Understand Cells Beyond Their Gene Panels
What if single-cell models learned what makes a cell itself rather than just predicting missing genes?
Most single-cell foundation models focus on masked gene reconstruction. This trains the model to fill in values rather than to learn meaningful cell-level representations, which is why they often underperform on downstream tasks such as annotation, transfer learning and spatial integration.
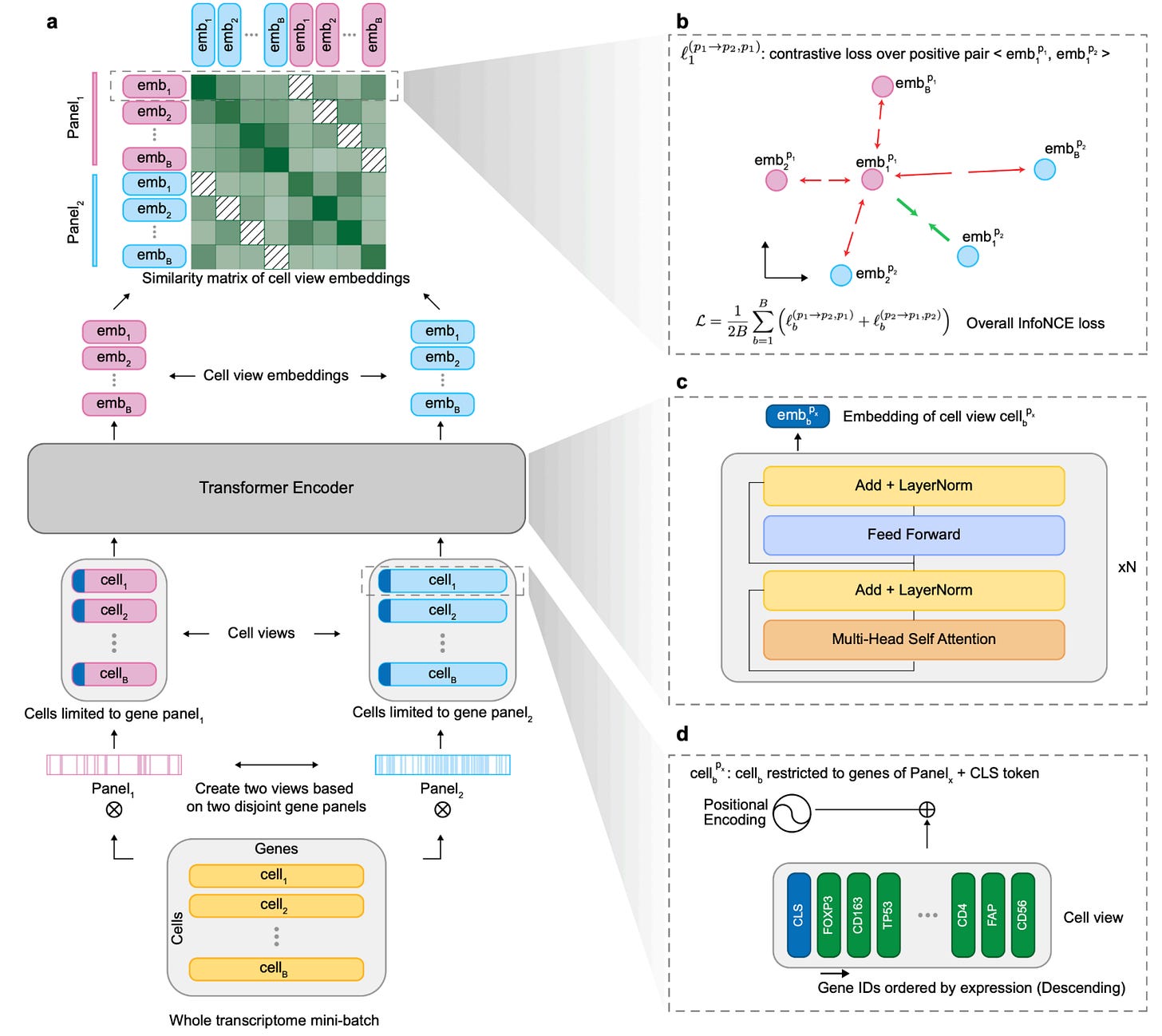
Researchers at Helmholtz Munich propose scConcept, a contrastive learning framework that teaches a model to recognise a cell from any subset of its genes. Each cell is split into two non-overlapping gene panels and the model learns to bring those representations together while pushing away other cells.
The result is a model that captures a technology-agnostic and panel-independent notion of cell identity.
🔬 Applications and Insights
1️⃣ Better cell-type annotation
Across bone marrow, brain and skeletal muscle datasets, scConcept outperformed existing foundation models by more than 5 per cent macro F1 and outperformed domain-specific tools such as CellTypist. A lightweight adapted version, scConcept+, improves results further with only minimal self-supervision.
2️⃣ Generalisation across technologies
Although trained only on dissociated scRNA seq, scConcept transfers naturally to spatial assays. In Alzheimer’s MERFISH datasets, it delivered the strongest zero-shot label transfer, and adaptation pushed performance beyond all baselines.
3️⃣ Stronger spatial gene imputation
scConcept and scConcept+ improved held-out gene prediction by 10 to 15 per cent compared with specialised approaches such as Tangram. Adaptation improved every gene rather than trading accuracy across transcriptome regions.
4️⃣ New capabilities reconstruction models cannot support
Because scConcept learns consistent structure from arbitrary gene subsets, it can:
perform mutual information based gene panel optimisation
map new spatial assays into existing atlases such as HLCA more accurately than scArches
💡 Why It Is Cool
scConcept shows that single-cell models do not need to predict genes to learn biology. By optimising directly for cell identity, it delivers robust, technology-independent embeddings and unlocks entirely new applications.
📄 Read the paper
⚙️ Code available on GitHub.
⚛️ Riff-Diff: One-Shot Enzyme Design That Rivals Directed Evolution
What if highly active enzymes could be designed in a single cycle rather than after years of evolution?
Most computational enzyme designs start 10⁵ to 10⁶ times slower than evolved enzymes and only become practical after screening thousands of variants.
Riff-Diff, developed by Graz University of Technology, achieves what previously required many rounds of directed evolution: the creation of efficient enzymes with fewer than 100 designed sequences per target.
Riff-Diff combines machine learning and atomistic modelling to scaffold catalytic arrays into de novo protein backbones. The system enforces precise fragment placement, deep substrate-shaped pocket formation and strict catalytic geometry. Catalytic residues are embedded into helical fragments with compatible rotamers, then RFdiffusion with custom potentials scaffolds these motifs into functional active sites.
🔬 Key Results and Applications
1️⃣ Retro-aldolase activity
More than 90 per cent of designs were active. The top variants achieved kcat values around 0.03 s⁻¹, delivering approximately a million-fold rate acceleration with strong stereoselectivity.
2️⃣ Morita-Baylis-Hillman reaction
More than 90 per cent of designs showed activity. The best catalyst outperformed a variant discovered only after screening more than 13,000 clones through directed evolution.
3️⃣ Structural accuracy
Crystal structures confirmed near-atomic precision. Catalytic side-chain RMSDs were generally below 1 Å.
4️⃣ Exceptional stability
Designs remained folded above 90°C and showed high chemical stability, consistent with predictions of well-packed cores.
💡 Why It Is Cool
Riff-Diff demonstrates that starting from well-characterised catalytic motifs and designing for structural precision enables true one-shot creation of efficient, stereoselective enzymes. It dramatically reduces reliance on long directed-evolution campaigns and accelerates catalytic discovery.
📄 Read the paper.
⚙️ Explore the code and datasets.
Thanks for reading Kiin Bio Weekly!
💬 Get involved
We’re always looking to grow our community. If you’d like to get involved, contribute ideas or share something you’re building, fill out this form or reach out to me directly.
Connect With Us
Have any questions or suggestions for a post? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website