⚛️ Nygen’s CyteType, Harvard’s ODesign, and Genentech’s GNEprop
Kiin Bio's Weekly Insights
Welcome back to your weekly dose of AI news for Life Science!
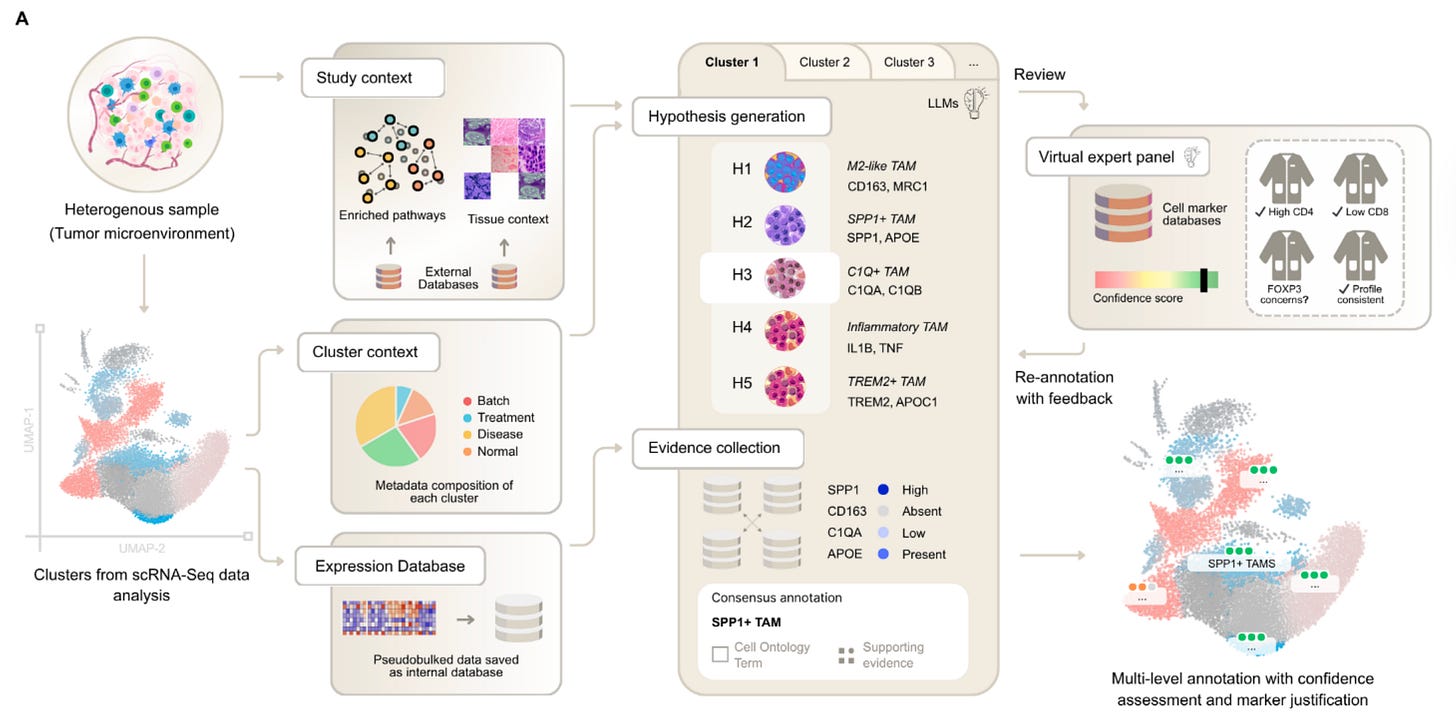
🧬 CyteType: Multi-Agent AI Transforms Cell Type Annotation
What if cell type annotation stopped being guesswork and became a transparent, evidence-driven process?
CyteType, developed by Nygen Analytics, is a multi-agent AI system that tackles one of the biggest bottlenecks in single-cell analysis: unreliable annotation in complex or disease contexts.
Where standard tools suffer accuracy drops of 15 to 30 per cent and provide little justification, CyteType generates competing hypotheses, validates them against external resources, and delivers calibrated confidence scores with full reasoning traces.
The system coordinates five specialised agents:
A Contextualiser that frames biological setting, an Annotator that tests hypotheses, a Reviewer that simulates expert consensus, a Literature Agent that connects to PubMed and ontologies, and a Summariser that consolidates everything into interpretable results.
Fully compatible with AnnData and Seurat, CyteType outputs Cell Ontology mapped labels and interactive HTML reports with embedded chat for iterative refinement.
🔬 Applications and Insights
1️⃣ Superior Performance
Across 205 clusters, CyteType outperformed reference-based tools with a 252 per cent increase in semantic similarity. It also outperformed GPT-based annotation by 389 per cent even when using identical models.
2️⃣ LLM Robustness
Evaluation across 16 language models showed performance remained stable and model agnostic. Closed-weight models such as GPT-5 and Claude Sonnet 4 achieved the highest accuracy. Open-weight models such as DeepSeek R1 and Qwen3 achieved around 95 per cent of peak performance at lower cost.
3️⃣ Trustworthy Predictions
Confidence scores correlated strongly with correctness (F = 23.88, p < 0.001). Heterogeneity flags detected mixed populations, and over 70 per cent of clusters showed consensus Cell Ontology labels across repeated runs.
4️⃣ Beyond Classification
Across 977 clusters, CyteType improved functional interpretation for 41 per cent, refined subtypes for 29 per cent, and triggered major reannotations for 30 per cent. In total it uncovered 327 Cell Ontology terms and 116 cell states.
💡 Why It’s Cool
CyteType turns annotation into a transparent and evidence-based process with quantifiable uncertainty.
Instead of guessing cell identities, it produces testable hypotheses grounded in literature, ontologies, and biological context.
📄 Check out the paper
💻 Try the code on GitHub.
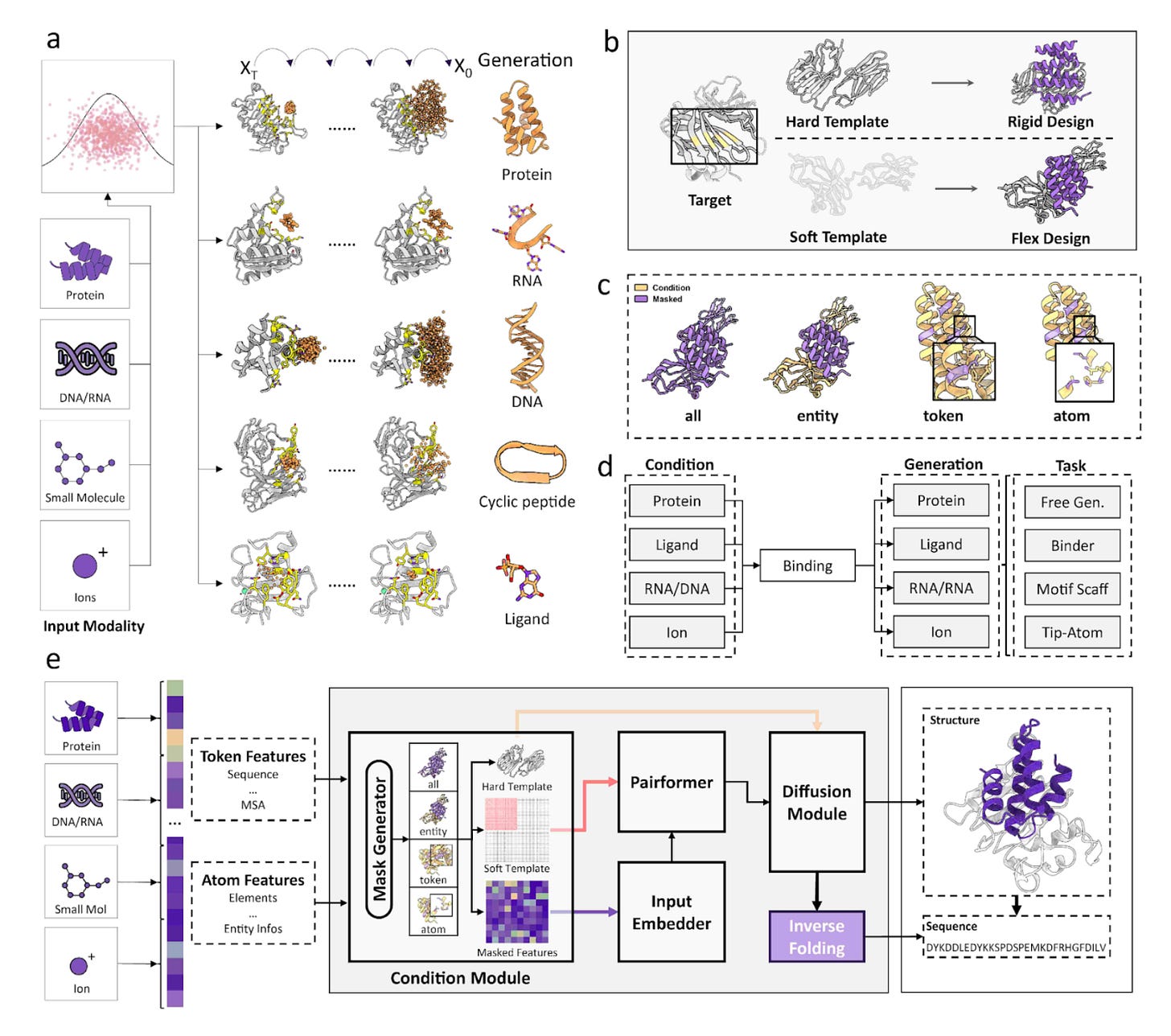
⚛️ ODesign: World Models Arrive for Biomolecular Design
What if one AI could design proteins, DNA, RNA, and ligands in the same model and understand how they interact?
ODesign is the first generative world model for biomolecular interactions. It unifies molecular design across proteins, nucleic acids, and ligands within a single framework.
Instead of relying on separate tools for each molecular modality, ODesign builds a shared token space that captures the physical and chemical principles underlying all-to-all interactions.
This is not incremental progress. It is a new way of designing biomolecules.
🔬 Applications and Insights
1️⃣ Unified Biomolecular Design
ODesign reasons jointly across proteins, nucleic acids, and small molecules using one architecture. This eliminates silos between modalities.
2️⃣ Physics-Informed Generative Process
The model incorporates physical constraints inspired by AlphaFold3, ensuring designs obey real molecular geometry and energetics.
3️⃣ Multi-Scale Control
Users can condition the model at any level, from global fold to individual atoms. This enables scaffold exploration, loop redesign, and precision active-site engineering.
4️⃣ True Cross-Modality Interactions
For the first time an AI model can design protein-binding DNA or RNA and nucleic-acid-binding ligands. These interactions were previously inaccessible.
💡 Why It’s Cool
ODesign ends the era of one-model-per-task molecular design.
It is fast, unified, physics aware, and accessible through a simple web interface.
It does not only generate molecules. It understands them.
📄 Read the paper.
⚙️ Try the code on GitHub.
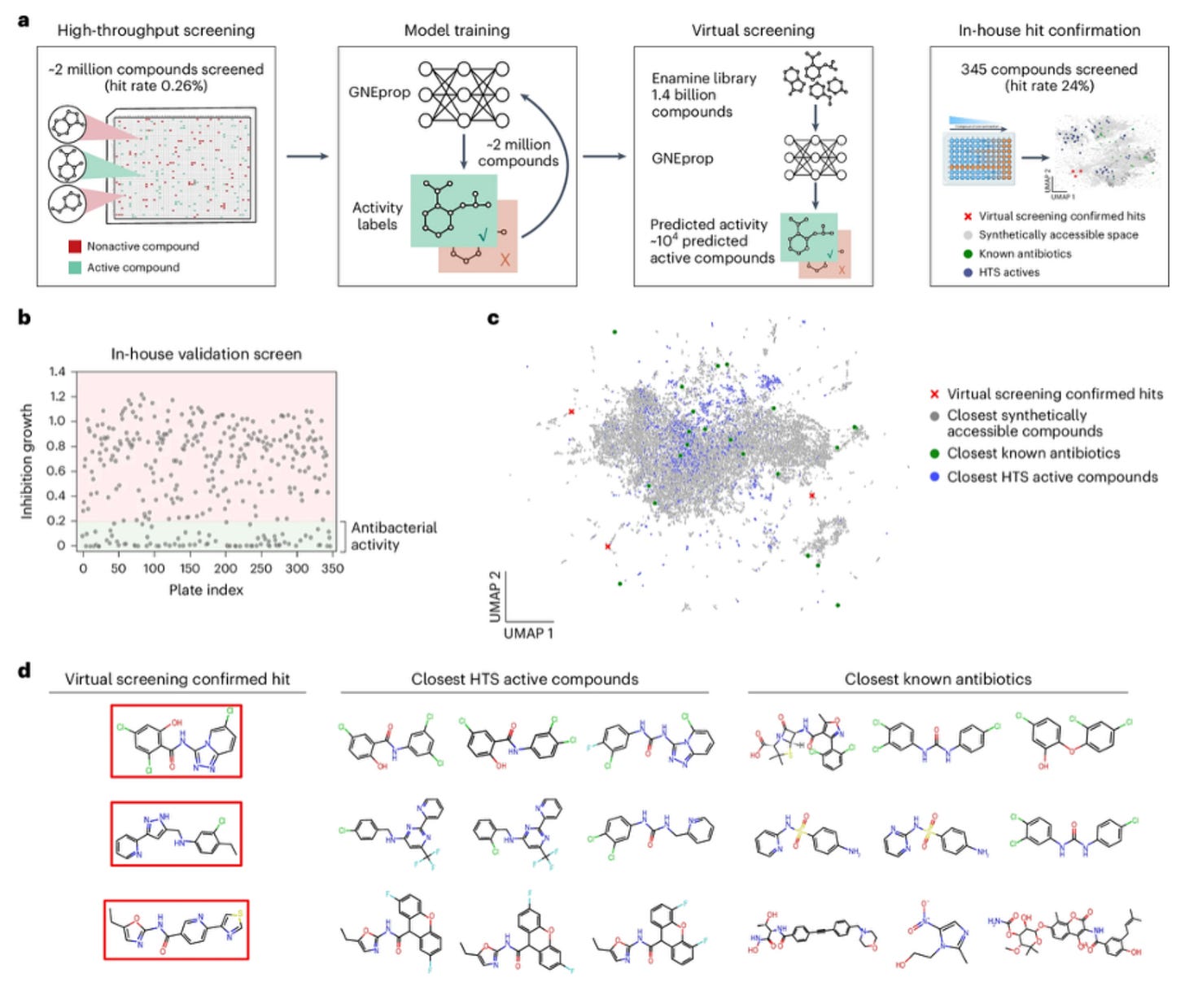
🦠 GNEprop: AI Unlocks Billion-Compound Search for New Antibiotics
What if antibiotic discovery could scale from testing millions of molecules to exploring billions?
GNEprop, developed by Genentech, combines high-throughput screening with deep learning to accelerate the search for new antibiotics. Antimicrobial resistance is projected to cause eight million deaths by 2050, and traditional discovery pipelines are not keeping pace.
Experimental screens usually cover one to ten million molecules. GNEprop learned from more than two million experimentally tested compounds and applied these insights to approximately 1.4 billion synthetically accessible virtual molecules. This enables the discovery of antibacterial candidates that would never appear in standard libraries.
🔬 Applications and Insights
1️⃣ Billion-Scale Virtual Screening
GNEprop enables virtual screening of billions of molecules, greatly expanding the chemical space that researchers can explore.
2️⃣ Ninety-Fold Improved Hit Rates
By prioritising the most promising candidates, GNEprop achieved an estimated ninety times improvement in confirmed hits compared with the initial experimental screen.
3️⃣ Breaking Chemical Library Bias
Traditional libraries favour known scaffolds. GNEprop generalises learned antimicrobial features to massive virtual libraries and identifies structurally novel compounds.
4️⃣ Integrated with Synthetic Libraries
The model connects directly to 1.4 billion synthesizable molecules, opening access to chemical space that has never been explored experimentally.
💡 Why It’s Cool
GNEprop identified 82 confirmed antibacterial hits from a billion-compound library. Many were structurally unprecedented and active against strains beyond the original training data.
📄 Read the paper.
⚙️ Try the code.
Thanks for reading Kiin Bio Weekly!
We’re always looking to grow our community. If you’d like to get involved, contribute ideas or share something you’re building, feel free to reach out.
Connect With Us
Any questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website