OpenFold3: Building the Next Generation of Open-Source Protein Structure Prediction
Deep Dive | Edition 3
Welcome back to the deep dive, where we break down the AI tools and data reshaping how new drugs are discovered. In each edition, we speak directly with the teams behind these tools to explain what they solve, how they work and where they are going next.
This edition focuses on OpenFold3 (OF3), a next-generation open-source model for protein structure and complex prediction. OF3 accurately predicts how a protein folds into a three-dimensional structure, using only the sequence of amino acids as input. We spoke with Woody Sherman, Chair of the OpenFold Consortium and CIO of Psivant Therapeutics, and Lukas Pluska, the Senior Commercial Operations Manager of Apheris, to understand how OF3 is being built, used, and fine-tuned by industry. The result is an open foundation model designed not just to predict protein structures, but to push forward drug discovery workflows.
🔴 The Problem
Protein structure prediction tools have come a long way since AlphaFold2, and models like AlphaFold3 are already delivering value for leading drug discovery companies but adoption across the broader field remains limited due to commercial constraints. While AlphaFold3 represents a major leap forward, it’s locked behind restrictions that prevent widespread access and real-world application in pharma pipelines.
This is where OpenFold3 changes the game. The goal of OF3 isn’t to reinvent the wheel. The first step is to match AlphaFold3’s capabilities and performance while providing a fully open-source foundation model that will spark innovation and improvements across the biotech and techbio ecosystems. As Woody Sherman, Chair of the OpenFold Consortium, put it: “Democratising state-of-the-art foundation models for biology is essential to unlocking their full potential in industry. By engaging a global community of researchers and practitioners across biotech, pharma, academia, and government labs, the OpenFold Consortium can focus on the most pressing real-world challenges and continue to raise the bar for the entire field. OpenFold3 is just the beginning of a new wave of AI tools in drug discovery. OpenFold3 is not the final solution, but it is a big step forward for the community and we look forward to building on capabilities such as binding affinity, folding stability, and generative modeling.”
Compounding this is a data problem. The Protein Data Bank contains over 200,000 structures, but the majority of protein-ligand complexes feature sugars, natural cofactors, metabolites or small peptides, not drug-like compounds. Meanwhile, pharma companies have thousands of proprietary complexes, with drug-like small molecules, but few mechanisms to train AI models on them while protecting intellectual property. It’s not all about having better data out of the box, although that does help. It’s also about giving researchers the freedom to make models better themselves through fine-tuning with proprietary data.
Now imagine fine-tuning OF3 not only on one company’s dataset, but across tens of thousands of proprietary structures from multiple pharma giants. This isn’t a hypothetical. It’s exactly what’s happening through the AI Structural Biology (AISB) Network, coordinated by Apheris. Companies like AbbVie, Johnson & Johnson, Bristol Myers Squibb, and Takeda are contributing their internal structural data via federated learning. Each keeps their IP locked down locally, but the model learns from all of it. The real breakthrough is fine-tuning across a collective pool of drug-relevant chemistry that no single organisation could assemble alone.
⚙️ What OF3 Does and Its Key Advantages
OpenFold3 is a deep learning model that predicts the 3D structure of protein-ligand, protein-protein, and antibody-antigen complexes directly from sequence and ligand input. It performs co-folding, meaning it folds all components jointly, rather than docking a ligand into a pre-folded protein. The model is inspired by AlphaFold3’s diffusion-based architecture and draws from prior work on AlphaFold-Multimer. It supports end-to-end prediction of complexes, including nucleic acids and small molecules.
Importantly, OF3 is engineered for portability. It’s written in open PyTorch, deployable via Docker containers, and supports batch inference for scaling in cloud or local environments.
Benchmarking of OF3 vs AlphaFold3
OF3 achieves comparable performance to AF3 on many co-folding tasks, including the PoseBusters and Runs’n’Poses benchmarks.
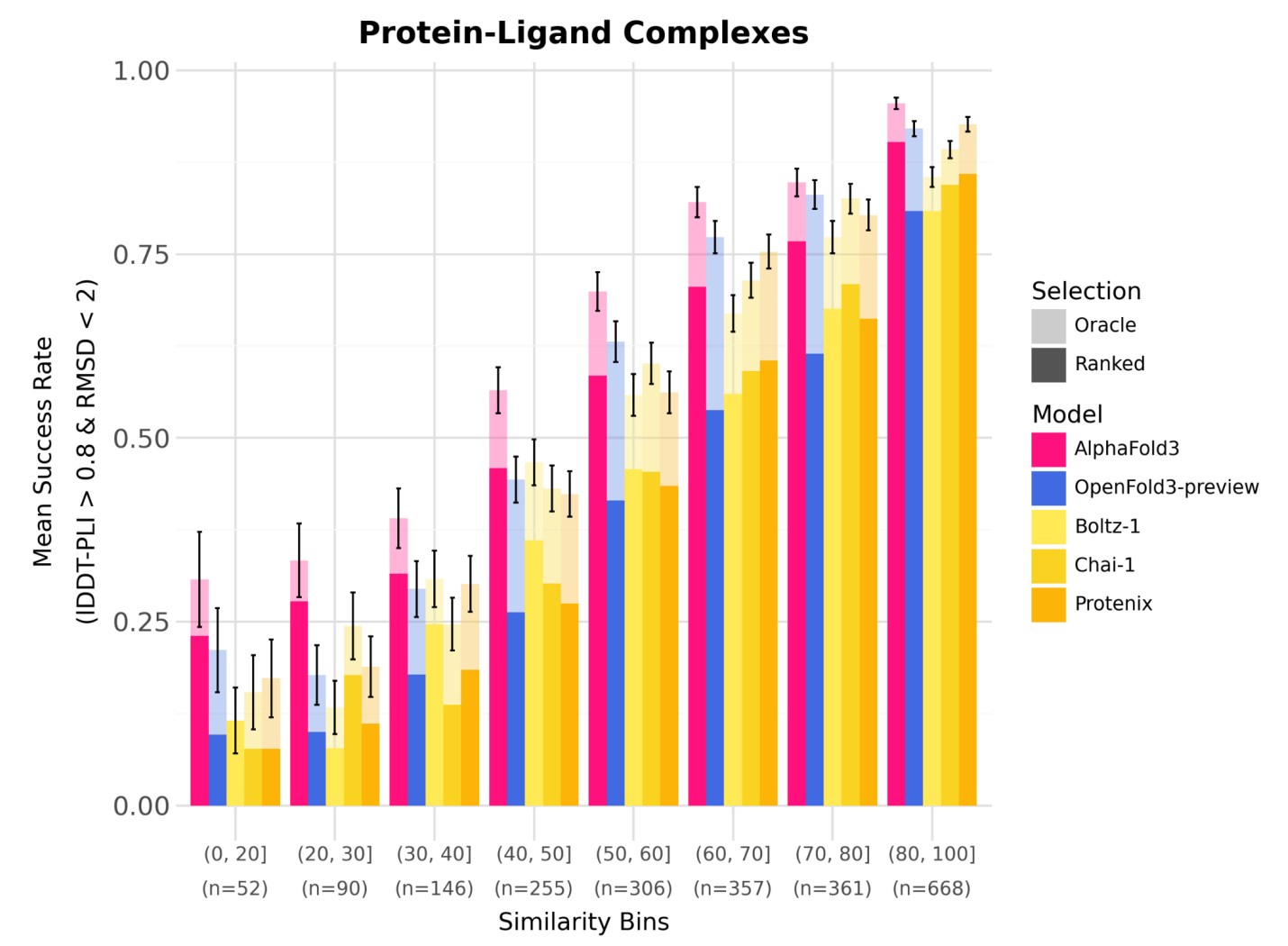
In some internal tests, especially on antibody-antigen and nucleic acid structures, OF3 has demonstrated better pose prediction than other open models like Boltz-2.
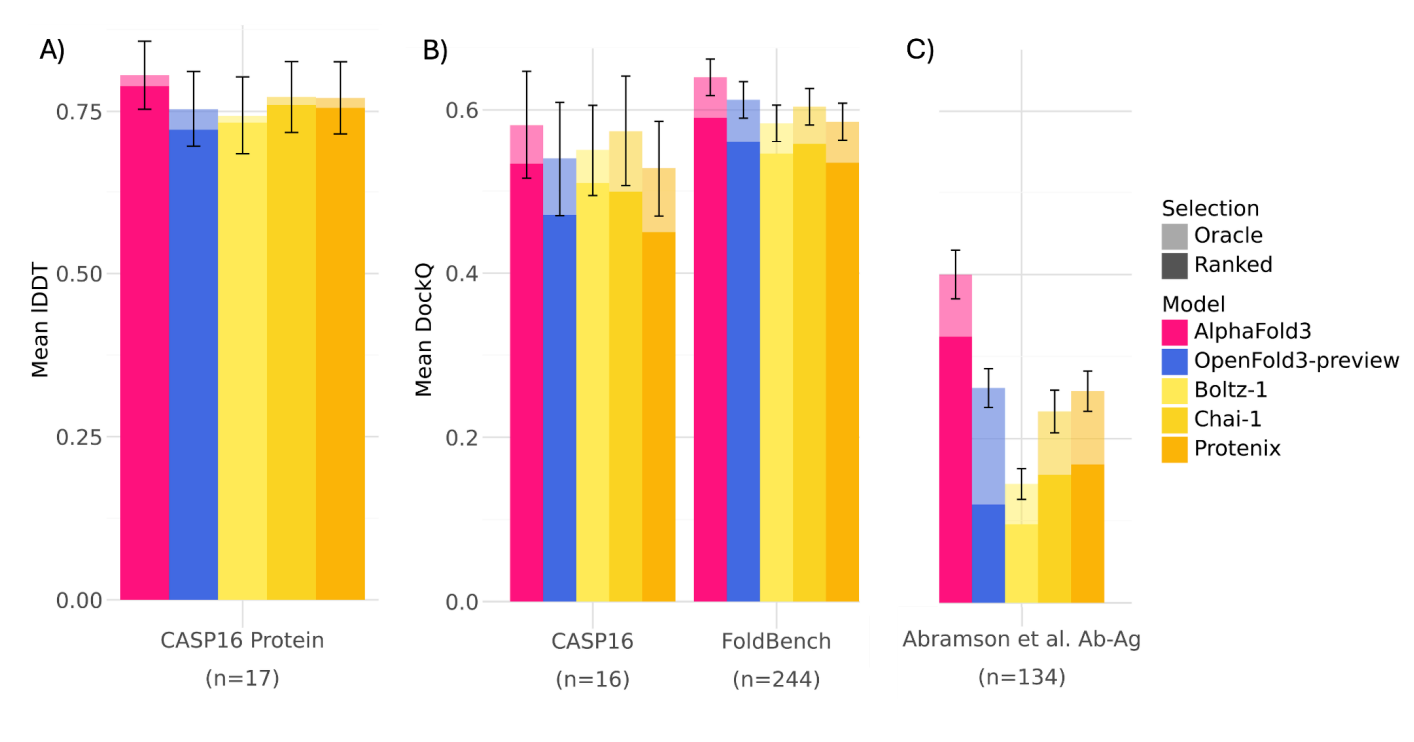
OF3 also benefits from inference-time scaling, using ensemble averaging and multiple poses to increase prediction robustness, similar to what’s observed in AF3.
Key Advantages of OF3
✅ Open License
Released under Apache 2.0, allowing unrestricted commercial and academic use, including fine-tuning and integration into proprietary workflows. For companies locked out of AlphaFold3 due to licensing restrictions, OF3 removes all barriers.🔁 Distillation Data
Trained with AlphaFold3-style pseudo-labels to expand learning beyond limited experimental structures.⚡ Inference-Time Scaling
Supports multiple sampled outputs per input complex for improved accuracy via ensembling.🧪 Rigorous Benchmarking
Evaluated using standardised protocols such as Runs’n’Poses, with careful controls to prevent training-test overlap.🏭 Industry Adoption
Already being adapted by early adopters including Novo Nordisk (drug discovery), Bayer Crop Science (trait design), and Outpace Bio (cell therapy engineering).🧰 Flexible Deployment
Available via NVIDIA NIM to get performance boost with GPU optimisation, along with ApherisFold.🌎 Ecosystem
A vibrant and diverse consortium spanning industries (biotech, pharma, crop science, and more) and modalities (antibodies, small molecules, peptides, and nucleic acids) to maximise longevity and practical impact.
💊 Importance and Application of OF3 in Drug Discovery
Protein-ligand structure prediction is essential in drug discovery, from early hit validation to structure-based lead optimisation but many AI models, especially those trained only on monomer structure, struggle when applied to drug-like small molecules or multi-chain protein complexes.
OF3 is purpose-built to handle:
Small molecule binding: for rapid generation of hypotheses, docking alternatives, and refinement inputs.
Antibody-antigen modelling: with dedicated attention to multi-chain assembly and pose plausibility.
Prediction where experimental structures don’t exist: enabling modelling of novel targets or complexes inaccessible to crystallography or cryo-EM
🧬 Data Used for OF3
Public Data (PDB)
OF3 is initially trained on public structural data: the Protein Data Bank (PDB), containing ~200,000 protein structures. However, only ~5-10% of these are protein-ligand complexes relevant to drug discovery. For fair benchmarking against AlphaFold3, OF3 uses the same training cutoff date (September 30, 2021), meaning both models draw from the same limited pool of public data, around 8,856 PDB protein-ligand complexes. This is why the federated learning approach becomes critical for moving beyond these constraints.
Distillation Data
OF3 expands its training beyond experimental structures by using distillation: predicted structures from high-performing models like AlphaFold3 serve as pseudo-labels for sequences that lack experimental data. This allows OF3 to learn from a much larger pool of examples and generalise beyond the limited set of known protein-ligand complexes in the PDB.
Federated Data: Apheris & AISB
Together with the AlQuraishi Lab at Columbia University the AI Structural Biology (AISB) Network addresses the gab of missing public data for model training. In the AISB Network’s Federated OpenFold3 Initiative pharma giants AbbVie, Astex, Bristol Myers Squibb, Johnson & Johnson, and Takeda are fine-tuning OpenFold3 on data from several thousand experimentally determined protein–small molecule structures, creating one of the most diverse datasets assembled for model training in drug discovery todate.
AbbVie alone contributed 9,000+ protein-ligand structures. According to Apheris the total dataset size is 5x more structural data that is directly relevant for industrial drug discovery compared to the original public training data. And the best thing, companies keep their data private but receive the collaboratively trained model.
In total, tens of thousands of proprietary structures were used in OF3 fine-tuning. To put that in perspective, this is comparable in scale to the subset of drug-like protein-ligand complexes available in the public PDB (~5,000 structures when excluding sugars, metabolites, and cofactors).
Companies keep their data private but receive access to the improved model.
Federation allows training of AI models on distributed data without the need to centralise it. Combined with additional governance, privacy, and security measures it’s possible to keep data IP fully protected while training an enhanced model that learns from all the distributed datasets. The increased quantity and diversity of those proprietary pharma datasets for model training leads to enhanced model generalisation on drug-like chemistry. While the base model will be released publicly, the collaboratively enhanced model trained on additional proprietary pharma data will only be released to participating AISB members.
OF3 is currently in active federated training through the AISB Network. A public version will be released, but only contributing partners get the fine-tuned model. New projects are opening soon, reach out to Lukas Pluska if interested.
The base OF3 model was trained on over 300,000 experimental structures and 13 million synthetic examples. The AISB federated initiative then fine-tunes this foundation model on an additional 10,000+ proprietary pharma structures, creating a specialised version for contributing partners while the public receives the base model.
🔮 Future
Binding Affinity Prediction
Initiatives like OpenBind (UK government) are already using OF3 as a starting point for large-scale model fine-tuning across new datasets.
The consortium is already exploring the next frontier: training OF3 variants to predict binding affinity, not just structure. Given that binding affinity correlates with structural accuracy, OF3 may be well-positioned to outperform other models like Boltz in this space.
Protein Stability
The OpenFold Consortium is working with Gabriel Rocklin at Northwestern University to build a mega-scale experimental dataset of protein folding stability that will be used to train OpenFold to predict folding stability.
Integration with Other Tools
As mentioned, OF3 is part of a broader open ecosystem at OMSF.
OpenADMET: Predict drug-likeness and pharmacokinetics
OpenFF / OpenFE: Physics-based force field development and free-energy simulation
OpenFold-SoloSeq: Integration with sequence-based LLMs for structure
Together, these tools form an open infrastructure stack for structure-guided drug discovery, from target to lead.
What’s Next for Federation
The AISB Network is currently fully enrolled for the OF3 training phase, but new federated initiatives are being scoped across a range of drug discovery applications.
If your organisation works on hard-to-model protein targets, contributes structural data, or wants to shape the direction of open AI in biotech, now is the time to explore participation.
OF3 is proof that federated collaboration can produce models that benefit the entire field, without requiring any one company to give up their competitive edge.
📄 Read the white paper!
⚙️ Access the model on Hugging Face.
⚙️ Access the model on Github.
✅ The OpenFold3-preview is accessible through the NVIDIA NIM too!
Thanks for reading!
Did you find this newsletter insightful? Share it with a colleague!
Subscribe now to stay at the forefront of AI in Life Science and keep up with this upcoming season of deep dives.
Connect With Us
Have questions or suggestions for our next deep dive? We’d love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website