Riverside's ParameterizANI, Owkin's OwkinZero and EPFL's De Novo Protein Switches
Kiin Bio's Weekly Insights
Welcome back to your weekly dose of AI news for Life Science!
Reminder: Our Tools Atlas database contains the codes for the open-source tools mentioned in this issue and all previous newsletters!
Is there any task that you find drains your time in the drug discovery process?
Let us know what it is and we’ll send you open source tools specific to that issue.
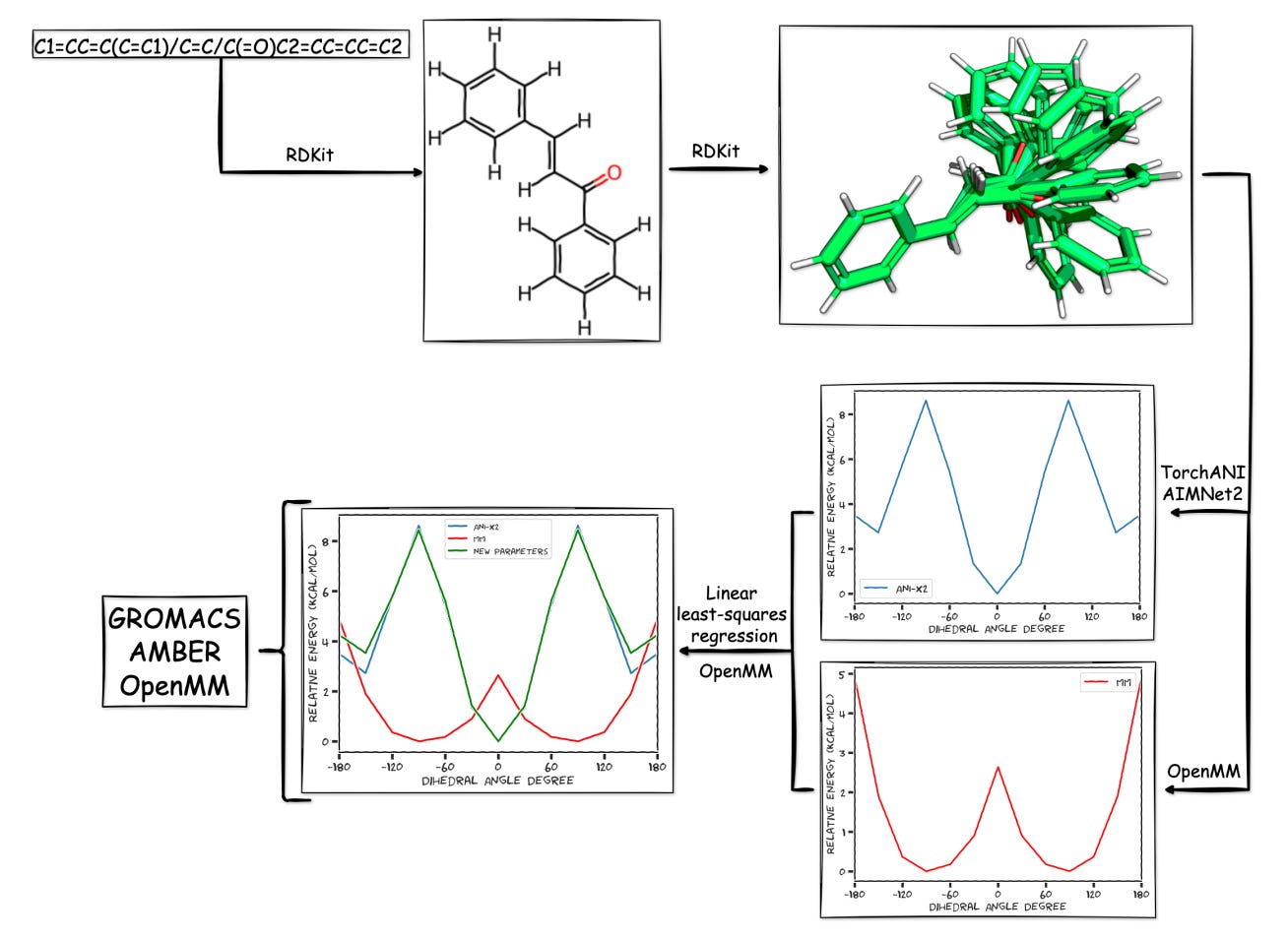
ParametrizANI: Fast, free, and accurate small-molecule parametrization with neural networks
Molecular simulations are only as good as the parameters you feed them.
But today, getting high-quality parameters for small molecules usually means wrestling with incredibly complex software stacks, expensive quantum calculations, and weeks of troubleshooting. For many labs, it is just not worth the effort.
ParametrizANI fixes that by making force field parametrization fast, free, and fully accessible.
Built by the University of California Riverside, this new tool uses neural network potentials and open-source chemistry frameworks to generate simulation-ready parameters directly from a SMILES string. It runs in Google Colab, requires no GPU access, and completes in under six minutes per molecule.
That means more students can learn molecular simulation, more labs can explore structure-activity relationships, and more pipelines can be automated at scale.
Applications and Insights
1. Fast and infrastructure-free
Parametrizes small molecules in under six minutes using only CPU resources in Colab. Makes high-quality molecular dynamics setup possible in environments without access to clusters or expensive software.
2. Quantum-informed accuracy
Matches HF/6-31G**-level dihedral energy profiles using ANI-2x potentials, providing reliable conformer energetics without running DFT calculations.
3. Flexible output formats
Exports topology and coordinate files compatible with OpenMM, GROMACS, AMBER, and Psi4. Supports GAFF and OpenFF for broad integration into downstream pipelines.
4. Customisable and transparent
Each step is implemented in a modular notebook with editable code. Users can change force field types, optimization methods, or add validation steps depending on their research needs.
The main takeaway from here is not just the level of speed or ease of use. It is the broad range of people this unlocks access for. ParametrizANI makes it realistic for any student, lab, or educator to run high-quality simulations without using expensive licenses or infrastructure. And because it is modular, users can customise every step of the process, from geometry optimisation to energy fitting.
I thought this was cool because it fixes one of the most painful bottlenecks in molecular modeling. ParametrizANI turns force field parametrization into something students can learn and labs can scale. And it does it with accuracy, transparency, and zero cost.
📄Check out the paper!
⚙️Try out the code.
OwkinZero: Teaching LLMs to Think Like Biologists
Most LLMs are great at complex maths and logical problems, but struggle with biological reasoning, the kind you need to link gene expression to drug response, or assess pathway activity across tissues.
OwkinZero changes that.
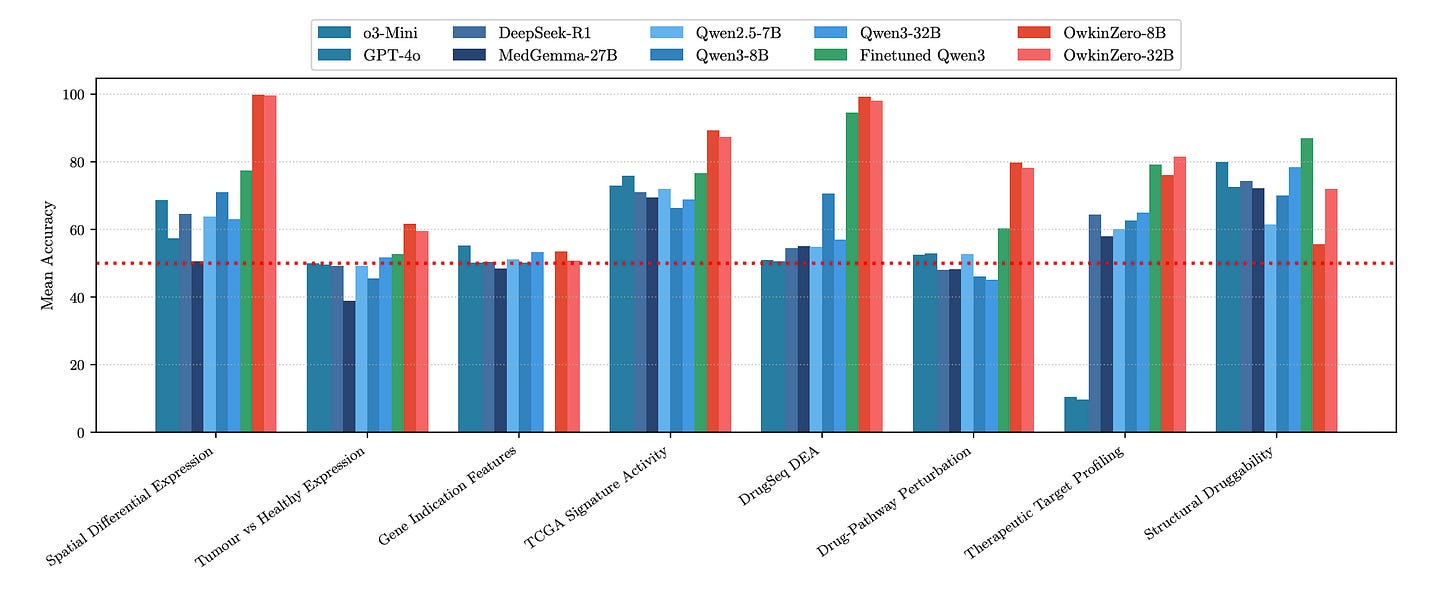
Built by OWKIN, it’s a family of 8 to 32B parameter models trained using Reinforcement Learning from Verifiable Rewards (RLVR) on over 300,000 structured Q&A pairs targeting real translational problems. The result? Stronger biological reasoning across expression data, drug effects, target modality, and structural druggability.
Think: Which cancer types share similar gene signature profiles? What pathways are most perturbed by CDK9 inhibition? Which of two binding pockets is more druggable?
These aren’t just high level trivia questions. They’re the kinds of decisions these researchers make every day. And surprisingly enough, even the best general-purpose LLMs (GPT-4o, MedGemma, DeepSeek-R1) fail to beat random guessing on most of them.
Applications and Insights
1. Benchmarks grounded in real biology
Eight datasets, 300K+ Q&A pairs covering expression, perturbation, structure, and modality. Each question is verifiable and requires reasoning, not recall.
2. Smaller beats bigger
OwkinZero-8B beats GPT-4o, MedGemma, and Qwen3-32B. For example, it scores 94.5% on drug perturbation (vs. 70.5% for Qwen3-8B) and 71.9% on structural druggability (vs. 60–64% for baselines).
3. Transferable reasoning
Specialist models trained on one task improved performance by up to +24 points on others. A model trained only on drug effects outperformed base models on spatial expression tasks.
4. Trade-offs in reasoning alignment
Mixture-trained models scored highest on accuracy but were less consistent in their explanations. Future work will likely require multi-stage fine-tuning to improve reasoning faithfulness.
I thought this was cool because it shows a realistic way forward for biology-focused models. Instead of pushing parameter count, OwkinZero focuses on data curation, smart task design, and domain-specific fine-tuning.
It also proves that biological reasoning isn't just one capability. It’s a layered set of skills: perturbation logic, expression interpretation, structural intuition. And those skills can be taught, transferred, and scaled with the right approach.
As the AI-for-biology space expands and develops with each day, this kind of focused infrastructure could become essential not just for answering questions, but for asking better ones too.
📄Check out the paper!
De novo protein optoswitches: Deep learning designs light-regulated dynamic proteins for synthetic biology
Designing Light-Controlled Proteins from Scratch
Most synthetic proteins are static. They do one thing, one way, at one time.
But we know well that natural proteins don’t work like that. They shift, fold, and respond to signals, turning on pathways, triggering functions, and regulating cells in real time.
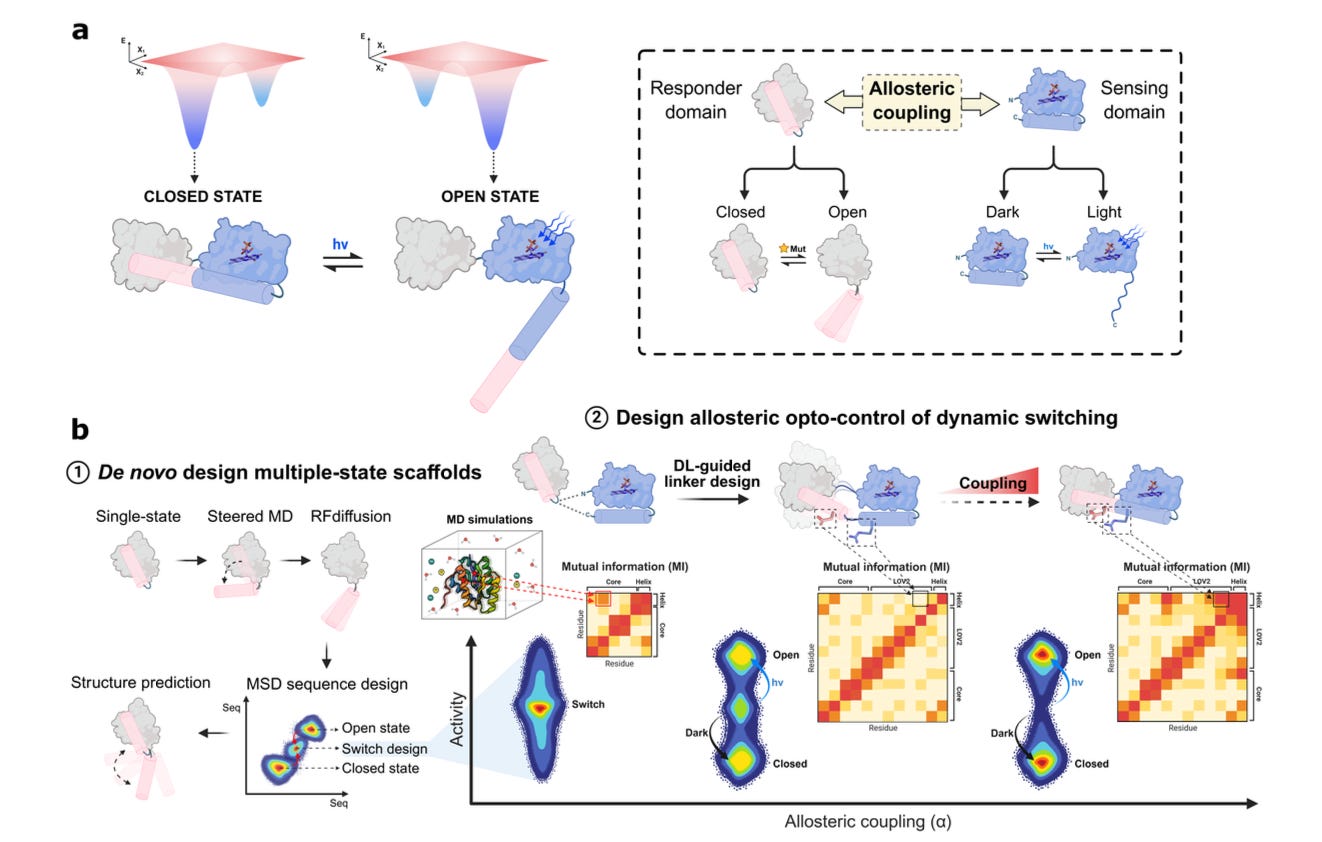
This new study from EPFL pushes synthetic design into that space. The team built de novo, light-controlled protein switches, so these are modular, reversible scaffolds that change shape and trigger function in response to blue light.
Everything is designed completely from scratch: the backbone, the switching mechanism, and the control logic. And its been shown to work in vitro, in cells, and even in living yeast.
Applications and Insights
1. Stable switching at atomic precision
Designed proteins toggle between open and closed states with under 2 Å RMSD to predicted structures. Both states are thermostable (Tm above 100 °C), with 15-24 targeted mutations controlling the conformational bias.
2. Strong, reversible light response
Fusing LOV2 domains via designed linkers triggered a 5× change in fluorescence on light exposure. The best variant showed R² = 0.94 in allosteric coupling and fully reversed within 10 minutes.
3. Programmable functions in cells
Adding motifs like NLS or α-pheromone let the switch control nuclear import (5.6× N:C ratio) and trigger light-dependent cell-cycle arrest in yeast, showing plug-and-play control of biological behaviour.
4. Deep learning-guided workflow
The designs combine RFdiffusion, ProteinMPNN, and AlphaFold2/3 with physics-based filtering. 27 out of 40 constructs folded correctly, and over half showed clear switching behaviour, which is a high hit rate for such complex design.
I thought this was cool because it makes proteins feel more like devices, not just structures but systems that do things. Turn on with light. Fold one way, then another. Trigger a function, then reset.
It’s the kind of work that shows what’s possible when you combine generative design with modular biology. You're not just engineering folds anymore. You’re engineering control.
And once you can control protein shape on command, you’re halfway to controlling cells, circuits, or even entire behaviours, using nothing but light. That’s the kind of foundation synthetic biology will be building on and it's exciting to think about what could come from this!
📄Check out the paper!
Thanks for reading!
Did you find this newsletter insightful? Share it with a colleague!
Subscribe Now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website