The Human Cytokine Dictionary, St Jude’s Combocat, and Barcelona’s NEMAT
Kiin Bio's Weekly Insights
Welcome back to your weekly dose of AI news for life science.
What’s your biggest time sink in the drug discovery process?
🧬 A Human Cytokine Dictionary at Single-Cell Resolution
What if cytokine biology were something we could read, not just observe?
Cytokines sit at the heart of immune signalling, yet their effects are notoriously hard to interpret. The same cytokine can trigger different responses depending on cell type, context and donor, and decades of studies have produced fragmented and often incomparable results.
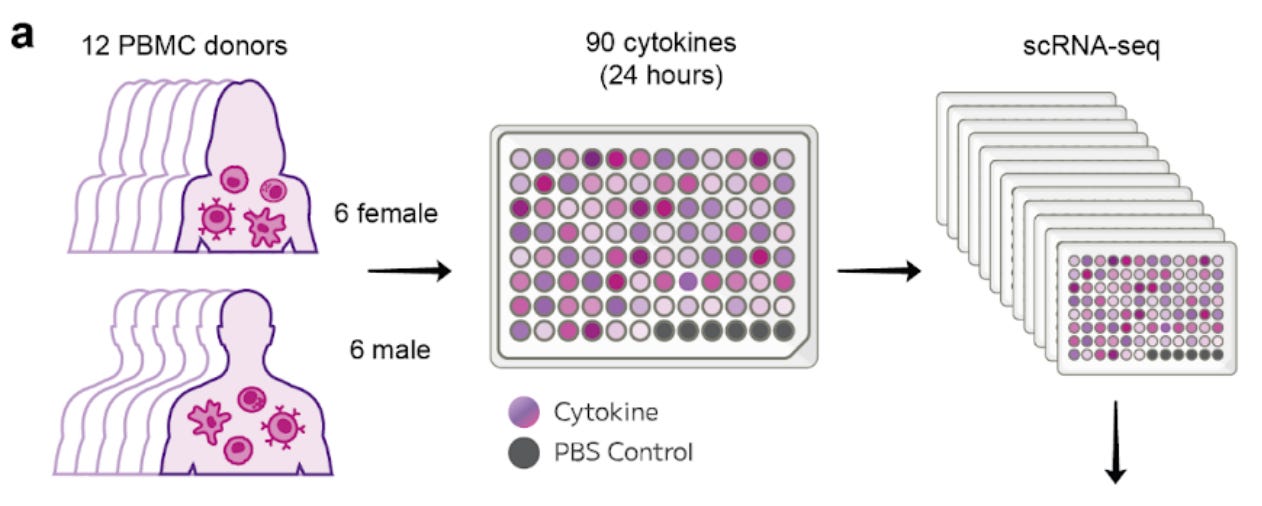
Researchers now present the Human Cytokine Dictionary, a systematic, cell type, resolved map of cytokine responses built from nearly 10 million single cells profiled across 90 cytokine perturbations in primary human PBMCs.
The result is not just a dataset, but a reference framework for decoding cytokine activity in health and disease.
🔬 Applications and Insights
1️⃣ A unified map of cytokine responses
The dictionary reveals which cytokines drive broad, pan-immune responses and which act in highly cell-specific ways. Signals such as IL-15 affect almost every immune cell, while others are sharply restricted to particular populations.
2️⃣ Donor variability without losing consensus
Despite clear donor-to-donor differences, often linked to baseline interferon states, most cytokine responses converge on robust and reproducible transcriptional programmes.
3️⃣ Cytokine-induced immune programmes
Rather than isolated gene lists, the study defines reusable immune programmes that capture functional states such as cytotoxic activation, antiviral defence and myeloid reprogramming.
4️⃣ Decoding disease and spatial biology
Using the accompanying huCIRA software, the dictionary is applied to lupus, multiple sclerosis and spatial cancer datasets, enabling inference of active cytokines even when cytokines themselves are not directly measured.
💡 Why It’s Cool
This work turns cytokine biology from anecdotes into infrastructure.
Instead of asking what a single cytokine does in isolation, researchers can identify active immune programmes, infer their drivers and understand how signals coordinate across cells. It provides an open, scalable and AI-ready foundation for predictive immunology and rational immunotherapy design.
📄 Read the paper
⚙️ Explore huCIRA
⚗️ Combocat: Making Drug Combination Discovery Practical Again
What if drug combination screening focused on better experiments rather than more of them?
Drug combinations underpin modern medicine, from oncology to infectious disease. But once more than one drug is involved, the experimental space explodes. Plates multiply, dose matrices grow, and meaningful coverage becomes infeasible.
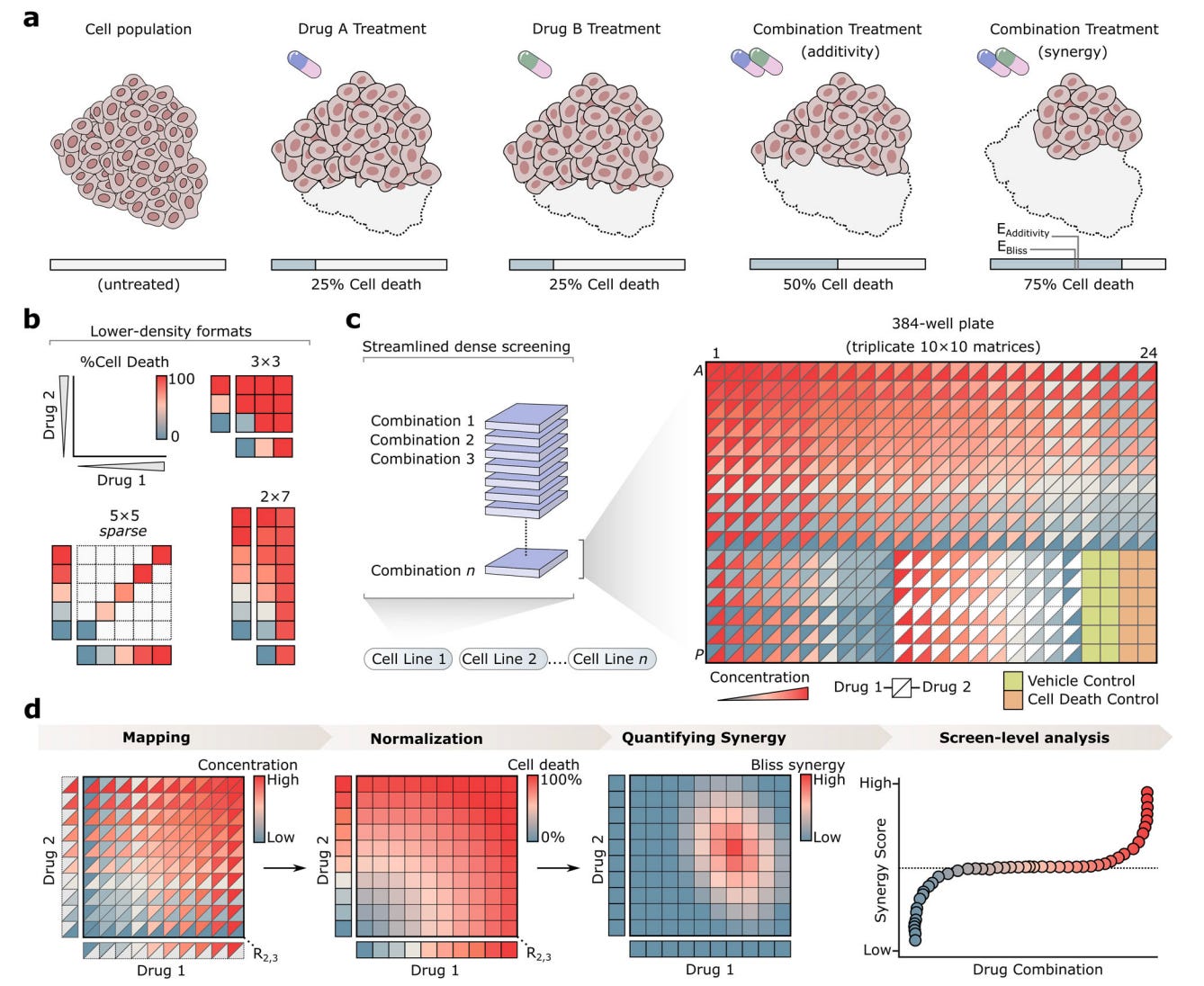
Researchers at St. Jude Children’s Research Hospital introduce Combocat, an open-source platform that integrates acoustic liquid handling with machine learning. It unifies wet-lab experimentation and computational inference into a single workflow designed to make large-scale combination screening practical.
This is not about running more experiments. It is about running better ones.
🔬 Applications and Insights
1️⃣ Dense, high-quality reference data
Combocat generates full 10 by 10 dose matrices, producing over 290,000 measurements across 806 drug pairs with strong assay quality, reflected in mean Z′ scores of around 0.75.
2️⃣ Sparse screening at real scale
By measuring only 10 dose pairs per combination, the platform reduces experimental effort by roughly 90 per cent and reconstructs the remaining response landscape computationally.
3️⃣ Machine learning that earns its keep
An ensemble of regression models predicts missing responses with a median R² of approximately 0.95, closely matching results from fully measured experiments.
4️⃣ Proof at meaningful scale
Using this approach, the team screened 9,045 drug combinations in a single neuroblastoma cell line, the largest such screen reported to date using modest resources.
💡 Why It’s Cool
Combocat reframes drug combination discovery as a design problem rather than a brute-force one.
It respects biological complexity without overwhelming researchers with plates and spreadsheets, and its open-source nature invites the community to build on it together.
📄 Read the paper
⚙️ Try the platform
🧪 NEMAT: Bringing Free-Energy Accuracy to Membrane Proteins
What if binding affinity predictions finally accounted for the membrane properly?
Membrane proteins are among the most important drug targets, yet predicting ligand binding remains difficult. Lipids complicate everything. Ligands partition into membranes, diffuse along them and only sometimes bind the protein. Traditional free-energy workflows struggle to disentangle these effects.
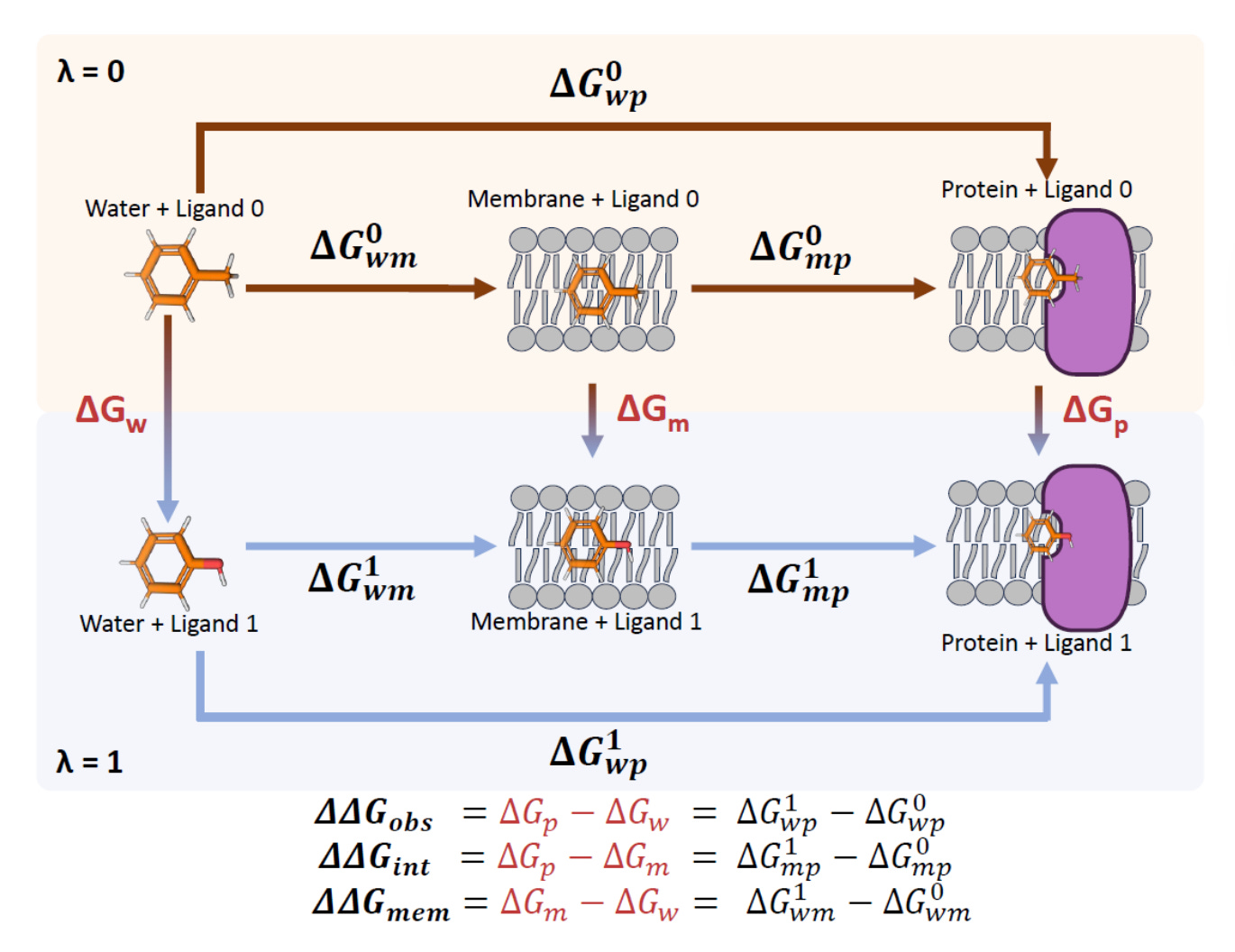
Researchers at IQAC-CSIC and the University of Barcelona introduce NEMAT, an open-source framework that applies non-equilibrium free-energy calculations to membrane systems in a way that is automated, reproducible and usable.
This is about making accuracy routine rather than heroic.
🔬 Applications and Insights
1️⃣ Membrane-aware binding predictions
NEMAT performs alchemical free-energy calculations in water, membrane and membrane-embedded protein systems, separating membrane partitioning from true protein binding.
2️⃣ Accuracy that holds up
On a benchmark of P2Y1 GPCR ligands, NEMAT reproduced experimental binding trends with an RMSE of around 1.7 kcal/mol, comparable to established equilibrium methods.
3️⃣ Non-equilibrium done properly
By controlling transition length, replica averaging and work distribution overlap, the pipeline delivers stable and reproducible free-energy estimates without extensive manual tuning.
4️⃣ Designed for real drug discovery
The workflow scales cleanly across GPUs and replicas, making it practical for exploring structure–activity relationships in membrane-exposed binding pockets.
💡 Why It’s Cool
NEMAT separates ligands that truly bind proteins from those that only look good because they like membranes.
That distinction matters for selectivity, safety and lead optimisation. It is not flashy, but it is foundational, and that is exactly why it matters.
📄 Read the paper
⚙️ Explore the code
Thanks for reading Kiin Bio Weekly!
💬 Get involved
We’re always looking to grow our community. If you’d like to get involved, contribute ideas or share something you’re building, fill out this form or reach out to me directly.
Connect With Us
Have any questions or suggestions for a post? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website