UC Berkeley’s NucleusDiff, Genentech’s Nona, and CeMM’s CellWhisperer
Kiin Bio's Weekly Insights
Welcome back to your weekly dose of AI news for Life Science!
What’s your biggest time sink in the drug discovery process?
⚛️ NucleusDiff: Solving Drug Design’s Atomic Collision Problem
What if AI-generated drug molecules could stop bumping into themselves?
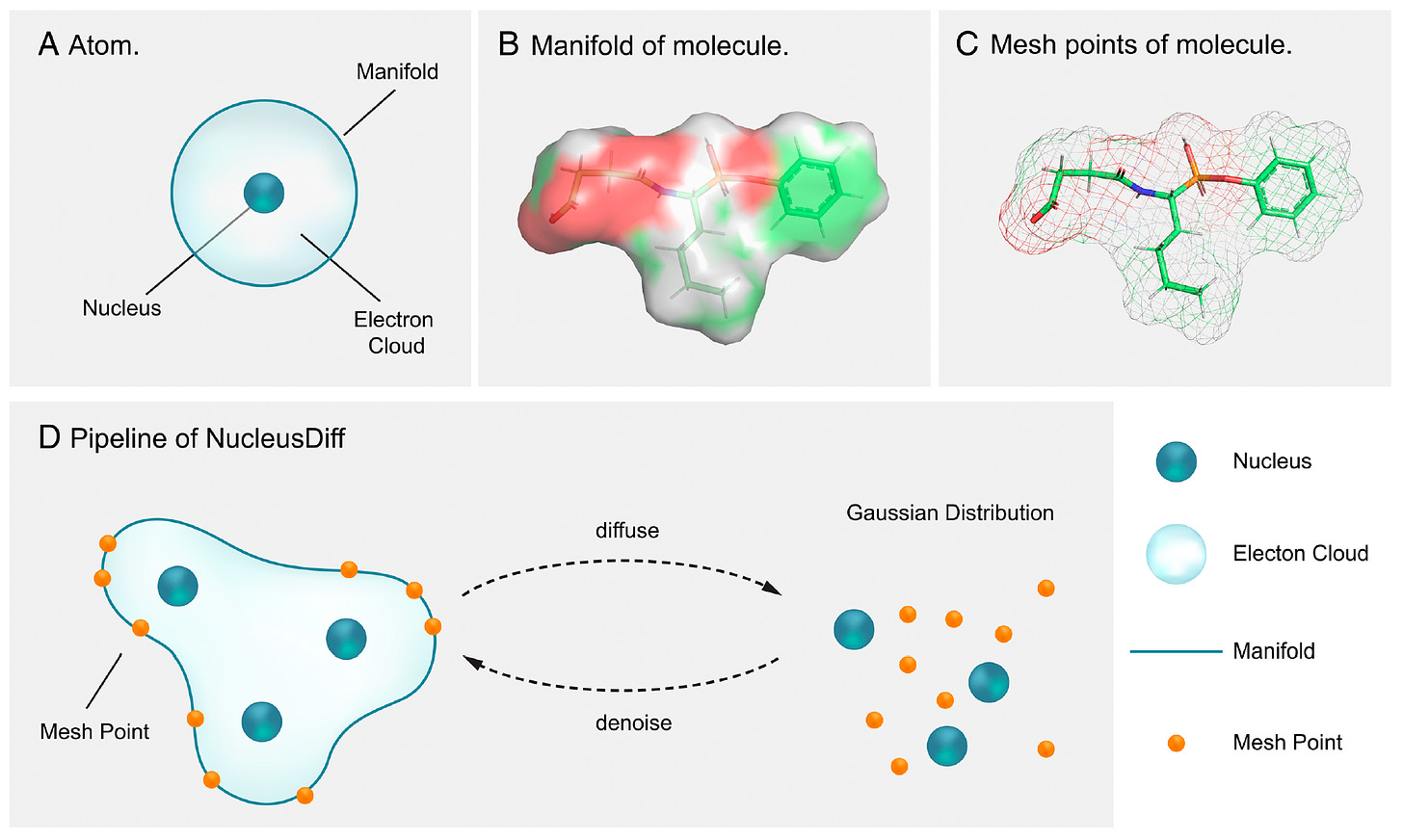
A team from UC Berkeley and Caltech have released NucleusDiff, a deep learning model that addresses a fundamental flaw in AI-driven drug design: atoms getting too close together.
While existing models generate molecules with impressive binding affinities, they often violate basic physics by placing atoms closer than their van der Waals radii allow.
NucleusDiff tackles structure-based drug design by modelling not just atomic nuclei positions, but also the spherical boundary around each atom. By discretising these boundaries into mesh points and enforcing distance constraints during training, the model generates physically plausible molecules that maintain proper atomic spacing while maximising binding affinity to target proteins.
🔬 Applications and Insights
1️⃣ Near-Perfect Collision Elimination
Testing on 100,000 protein-ligand complexes, NucleusDiff reduced atomic collision rates by up to 100%. The pairwise collision ratio dropped to essentially zero by the final sampling step.
2️⃣ Superior Binding Affinity
Achieved a 22.16% improvement in binding affinity over the previous best model, generating 60.1% high-affinity ligands compared with 56.3% for the baseline.
3️⃣ COVID-19 Success
Targeting the 3CL protease, NucleusDiff improved binding affinity by 21.37% and reduced collision rates by 66.67%, demonstrating strong real-world applicability.
4️⃣ Efficient Physics
The approach scales linearly with atoms rather than quadratically, making it both accurate and efficient for realistic drug candidates.
💡 Why It’s Cool
NucleusDiff shows how incorporating fundamental physical constraints into generative AI can improve both realism and performance.
By respecting the actual space atoms occupy, it bridges computational efficiency and chemical accuracy, producing drug candidates that are both potent and physically synthesisable.
📄 Check out the paper!
⚙️ Try out the code.
🧬 Nona: One Framework to Rule All Genomic AI Models
What if predicting, interpreting, and designing DNA could happen in a single model?
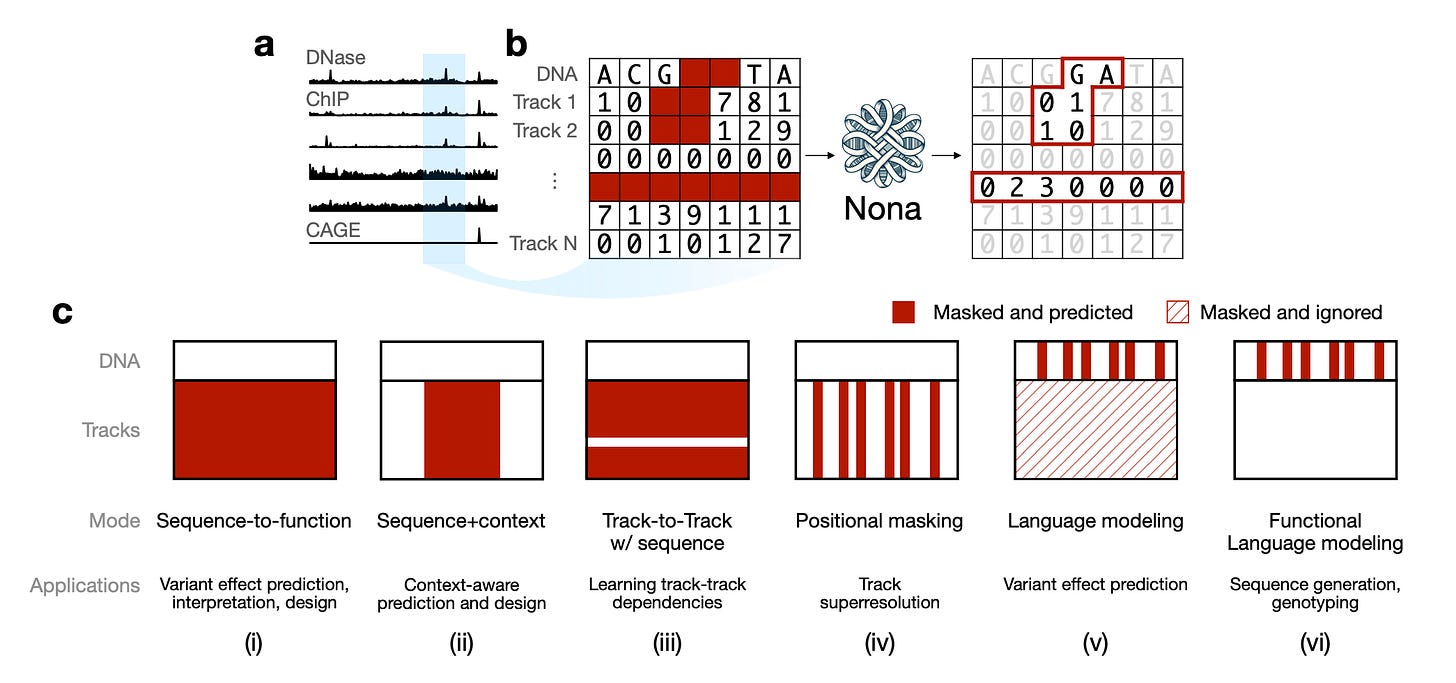
Researchers at Genentech have released Nona, a multimodal masked modelling framework that unifies three previously separate paradigms in functional genomics: sequence-to-function prediction, DNA language modelling, and generative design.
By treating masking as a universal interface, Nona eliminates the fragmentation that has long affected genomics AI.
The same architecture can perform radically different tasks without any code changes, simply by changing which parts of DNA sequence and functional data are masked.
Nona operates on both DNA sequence and base-resolution functional genomics data (such as DNase-seq and CAGE-seq), predicting masked values from unmasked context. Different masking patterns enable everything from variant effect prediction to regulatory element design.
🔬 Applications and Insights
1️⃣ Context-Aware Predictions
By conditioning on experimental measurements from adjacent genomic regions, Nona improved local functional predictions by up to 13%, correcting errors in heterochromatin and repeat-rich regions.
2️⃣ Functional Language Modelling
Unlike traditional DNA language models that learn repetitive sequences, Nona’s functional grounding emphasised transcription factor binding motifs. It also generates realistic regulatory sequences 16 times faster than previous methods.
3️⃣ Privacy Vulnerability Discovery
A small Nona model revealed an unrecognised security flaw: ATAC-seq fragment files leaked genetic information with 100% re-identification accuracy, correctly matching 83 individuals to their genotypes.
4️⃣ Universal Interface
The framework scales from compact 128bp models to large 196kb predictors, all sharing the same architecture. Different tasks are simply different masking patterns.
💡 Why It’s Cool
Nona shows how a single unified framework can replace an entire ecosystem of specialised tools.
It accelerates experimentation while revealing unexpected applications such as privacy vulnerabilities hidden in biological data.
📄 Check out the paper!
⚙️ They are working on releasing the code!
💬 CellWhisperer: Chat Your Way Through Single-Cell Data
Analysing single-cell RNA sequencing data just became as simple as having a conversation.
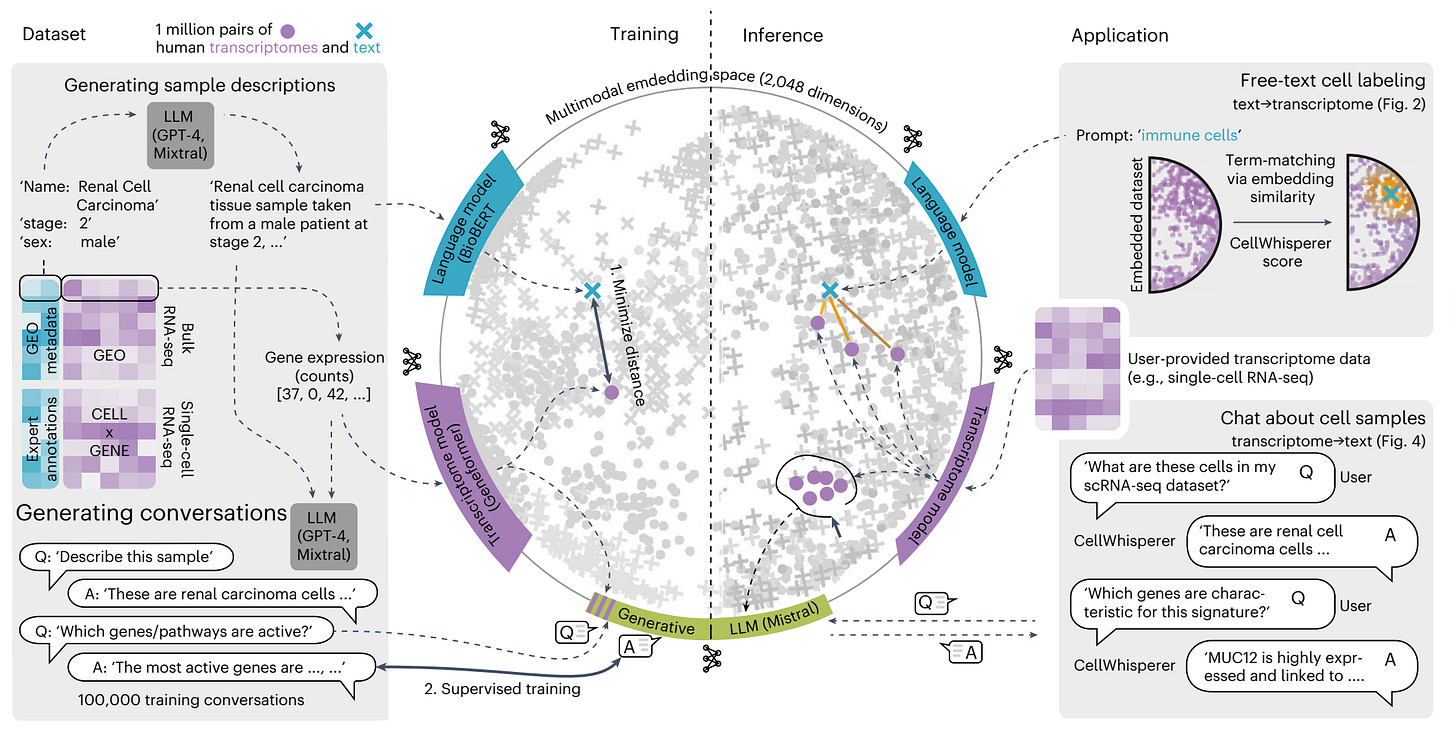
Developed by CeMM and collaborators, CellWhisperer is a multimodal AI that lets researchers explore gene expression through natural language.
Instead of writing code, users can ask questions such as “What are these cells?” or “Show me tissue-resident T cells in the intestine” and receive fast, biologically informed responses.
The system combines two models: an embedding model trained on over a million transcriptomes and a chat model adapted to interpret user queries in the context of uploaded gene expression data.
Integrated directly into the CELLxGENE browser, CellWhisperer enables seamless switching between visual exploration and interactive chat, helping users search, annotate, and reason about cells within one interface.
🔬 Applications and Insights
1️⃣ Zero-Shot Cell Type Prediction
CellWhisperer achieved 94% AUROC distinguishing 20 common cell types in the Tabula Sapiens dataset without any task-specific fine-tuning.
2️⃣ Organ Development Discovery
Applied to human embryonic development data, it identified marker genes for ten organs using simple text queries, uncovering at least ten new markers per organ supported by literature co-mentions.
3️⃣ Interactive Data Exploration
In a study of inflammatory bowel disease, CellWhisperer rapidly identified LGR5-expressing epithelial stem cells and revealed their depletion in inflamed tissue, an analysis that would normally require 400 lines of custom code.
4️⃣ Community-Scale Training
Trained on data from over 20,000 studies spanning diverse biological contexts, it provides broad coverage of human biology while maintaining cell-type specific resolution.
💡 Why It’s Cool
CellWhisperer turns single-cell exploration into a conversation.
It complements traditional bioinformatics while dramatically lowering the barrier to entry for researchers without programming expertise.
📄 Check out the paper!
💻Try the web app with public datasets.
⚙️ Try out the code.
Thanks for reading!
Did you find this newsletter insightful? Share it with a colleague!
Subscribe now to stay at the forefront of AI in Life Science.
Connect With Us
Have questions or suggestions? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website