UHN’s ECG-FM, Lorraine’s DynamicGT, and Weill Cornell’s HTS-Oracle
Kiin Bio's Weekly Insights
Welcome back to your weekly dose of AI news for life science!
What’s your biggest time sink in the drug discovery process?
🫀 ECG-FM: A Foundation Model for ECG Interpretation and Heart Failure Screening
What if a single foundation model could handle ECG interpretation and heart failure screening at once?
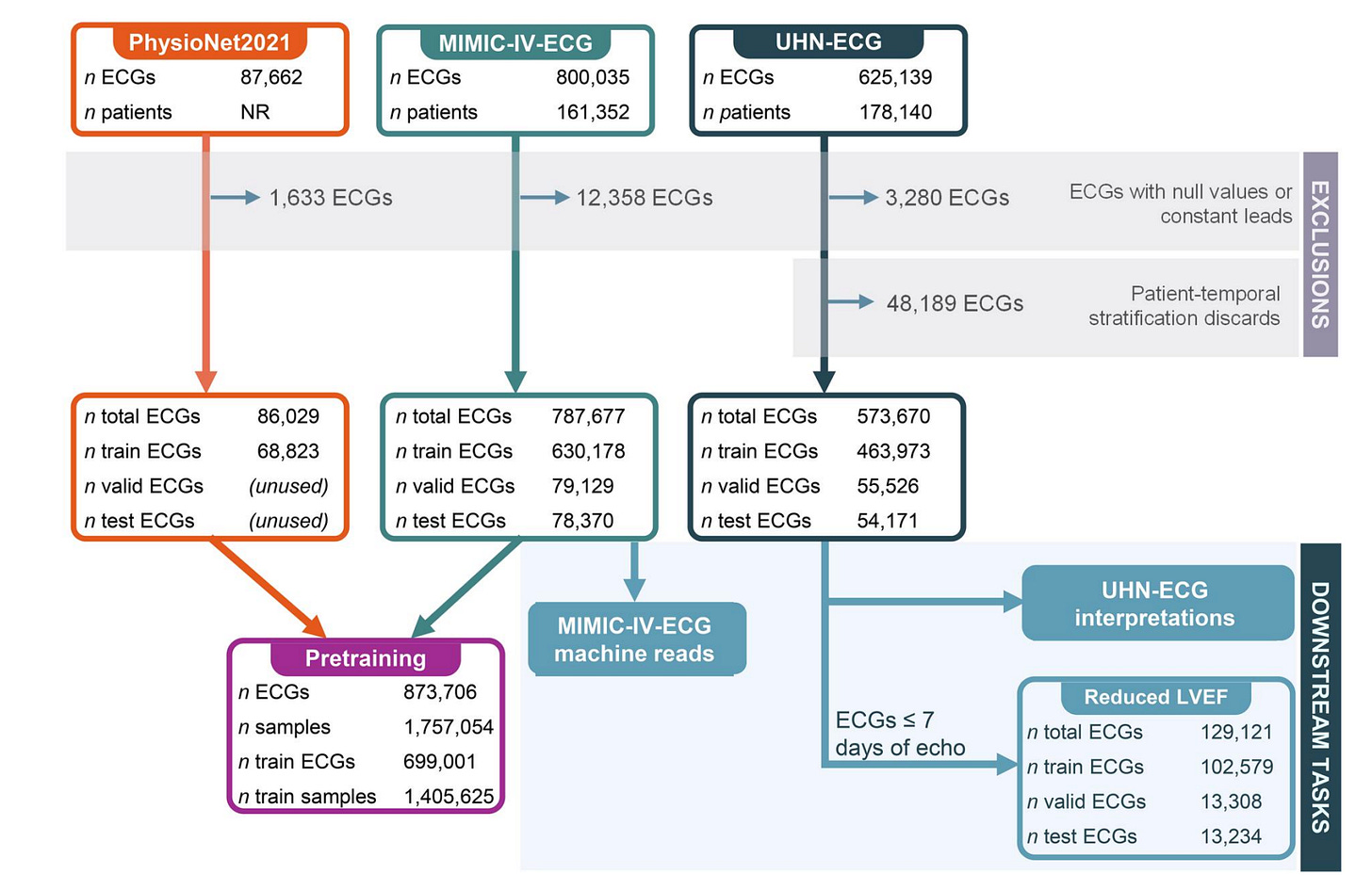
That is the goal of ECG-FM, a transformer-based foundation model for electrocardiograms developed by teams at University Health Network, the Vector Institute and the University of Toronto.
ECG-FM is trained on 1.5 million ECGs from public datasets, using no private clinical data. It combines masked prediction, contrastive learning and ECG-specific data augmentation to learn generalisable cardiac representations.
Crucially, the entire model is open-source, including weights, code and benchmark tasks, making it a public resource rather than a closed clinical system.
🔬 Applications and Insights
1️⃣ Label-efficient ECG interpretation
Using just 1 per cent of labelled training data, ECG-FM outperformed ResNet-based baselines across UHN and MIMIC tasks. It achieved an AUROC of 0.996 for atrial fibrillation and 0.929 for detecting LVEF ≤ 40 per cent.
2️⃣ Pretrained features that actually transfer
Linear probing on frozen embeddings achieved near-full performance on reduced LVEF and arrhythmia detection tasks, with no fine-tuning required.
3️⃣ Meaningful cardiac representations
Latent space clusters aligned naturally with rhythm, rate and pathology. UMAP visualisations separated sinus rhythm, bradycardia, tachycardia and conduction defects even without labels.
4️⃣ Robust to real-world noise
Up to 12.4 per cent of test ECGs were low quality. ECG-FM maintained strong performance, demonstrating resilience to artefacts common in clinical recordings.
💡 Why It’s Cool
ECG-FM is more than a strong model. It is an open, auditable foundation that anyone can adapt and build on. In a field dominated by siloed and opaque systems, that level of transparency matters.
📄 Read the paper
⚙️ Explore the code
🧬 DynamicGT: Predicting Protein Interfaces That Refuse to Sit Still
What if binding site prediction treated protein motion as signal rather than noise?
Most interface prediction models assume proteins are static and well folded. In reality, many interactions involve flexible and intrinsically disordered regions, where structure is transient and traditional predictors struggle.
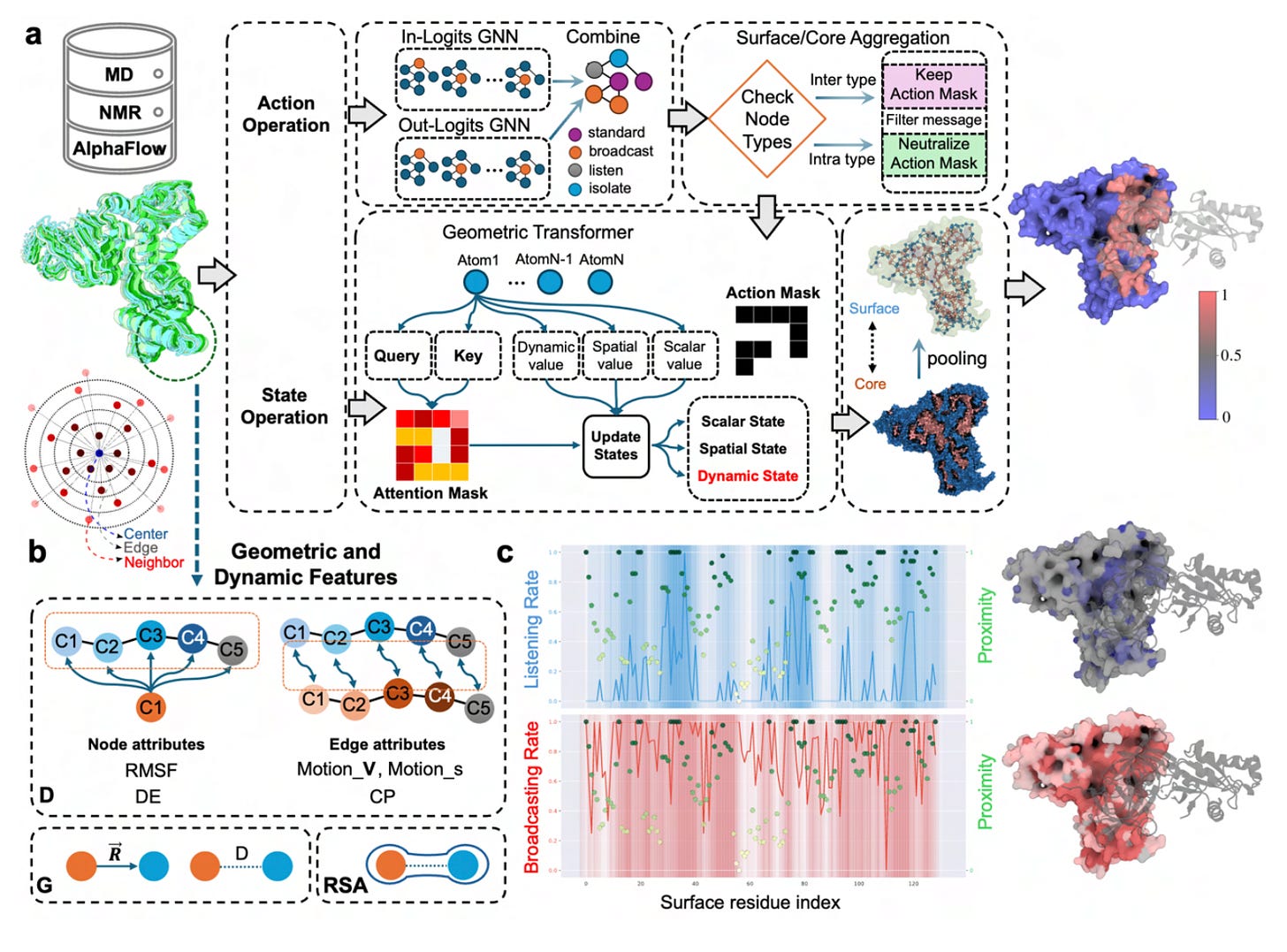
DynamicGT, developed by researchers at Université de Lorraine, CNRS, Inria (LORIA) and Université Grenoble Alpes, addresses this directly. Instead of relying on a single structure, DynamicGT learns from conformational ensembles, integrating molecular dynamics simulations, NMR structures and AlphaFlow-generated conformations.
The model predicts binding interfaces in contexts where flexibility and disorder dominate.
🔬 Applications and Insights
1️⃣ Interface prediction in disordered regions
Across benchmarks focused on intrinsically disordered proteins, including MFIB, Test42, FuzDB and IDRBind, DynamicGT achieved median ROC-AUCs up to 0.88, outperforming static predictors.
2️⃣ Learning from motion, not just shape
Dynamic features such as RMSF, directional entropy and inter-residue motion correlations significantly improved performance. Removing these features reduced ROC-AUC by around 3 per cent.
3️⃣ Strong performance without bound structures
Even with single-chain inputs and no complex structures, DynamicGT maintained ROC-AUC values around 0.85 using AlphaFlow-generated ensembles.
4️⃣ Improved spatial reasoning with GeoLoss
A distance-aware loss function penalised false positives based on spatial proximity, improving patch-level interface prediction rather than treating residues independently.
💡 Why It’s Cool
DynamicGT does not fight protein flexibility. It embraces it. By modelling motion directly, it finally gives intrinsically disordered regions the attention they deserve in interaction prediction.
📄 Read the preprint
⚙️ Try the code
🎯 HTS-Oracle: Smarter High-Throughput Screening for Difficult Targets
What if high-throughput screening tested the right compounds first?
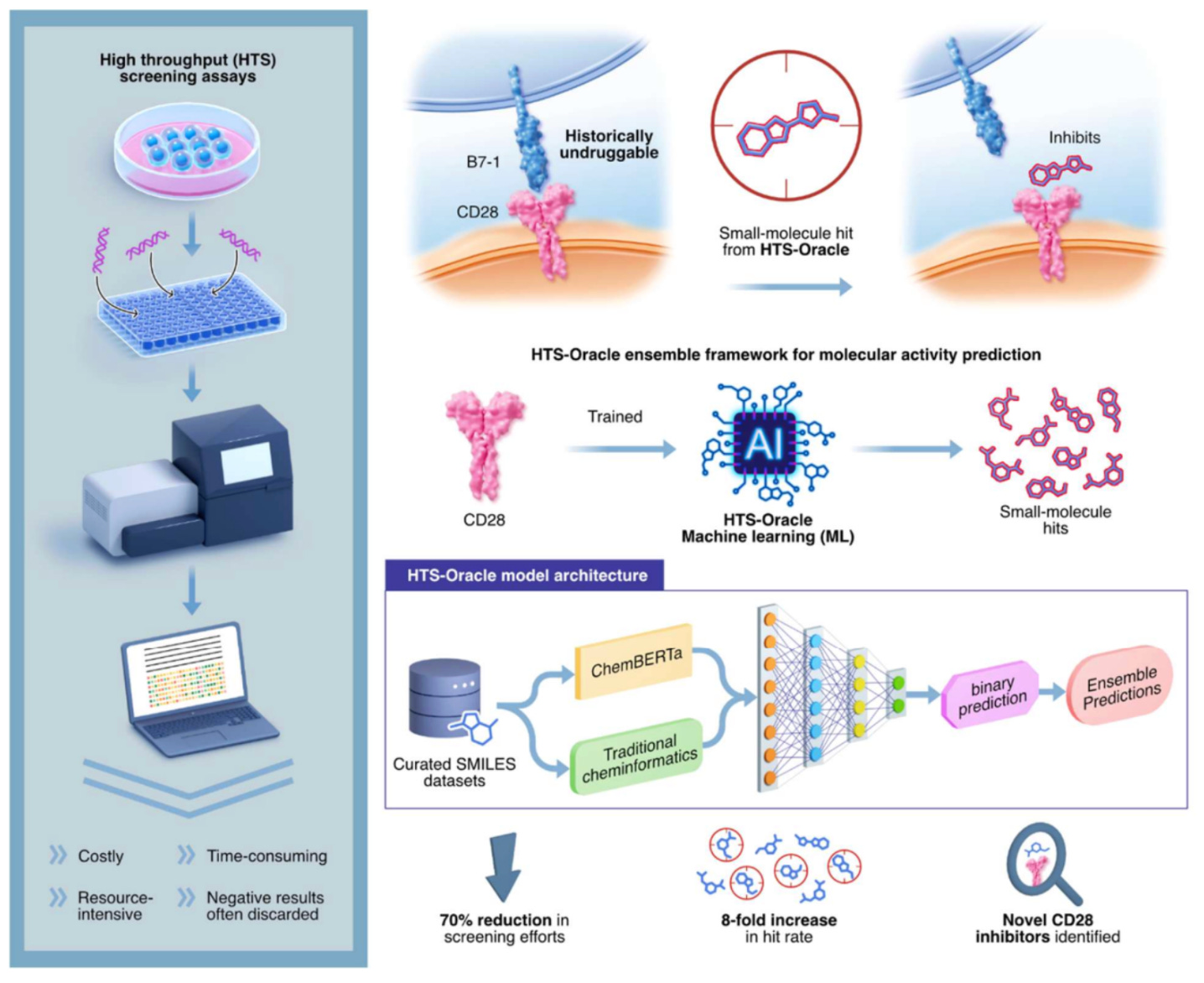
High-throughput screening is essential in early drug discovery, but it is expensive and typically produces very low hit rates, especially for immune targets that are considered difficult to drug.
HTS-Oracle, developed at Weill Cornell Medicine, is an AI-powered platform designed to prioritise compounds most likely to bind before experiments begin. It combines traditional cheminformatics with transformer-based molecular embeddings to capture complex structure–activity relationships.
The result is a more intelligent way to explore chemical space and dramatically improve hit rates.
🔬 Applications and Insights
1️⃣ Working with difficult immune targets
HTS-Oracle remained predictive even when active data were scarce. For CD28, where fewer than 2 per cent of compounds were active, the platform achieved an eight-fold increase in hits. It was also applied successfully to targets such as TREM2 and CHI3L1.
2️⃣ Substantially improved hit rates
From a 1,120 compound library screened against CD28, the model prioritised 345 compounds and achieved an 8.4 per cent hit rate, compared with typical rates below 2 per cent in conventional HTS.
3️⃣ Identification of biologically meaningful binders
Of the prioritised compounds, 29 were experimentally confirmed as hits. Two disrupted the CD28–B7.1 protein–protein interaction, a key signal in T-cell activation.
4️⃣ Retrainable by design
HTS-Oracle is built to incorporate new experimental data as it becomes available, allowing performance to improve with each screening cycle.
💡 Why It’s Cool
HTS-Oracle shows how AI can make early drug discovery more efficient, even for targets long considered undetectable. By reshaping what gets tested first, it reduces cost, accelerates learning and makes screening campaigns far more informative.
📄 Read the paper
⚙️ Try the code
Thanks for reading Kiin Bio Weekly!
💬 Get involved
We’re always looking to grow our community. If you’d like to get involved, contribute ideas or share something you’re building, fill out this form or reach out to me directly.
Connect With Us
Have any questions or suggestions for a post? We'd love to hear from you!
📧 Email Us | 📲 Follow on LinkedIn | 🌐 Visit Our Website