What task in your drug discovery work is the most time consuming?
We will send you open-source tools relevant to your pain point.
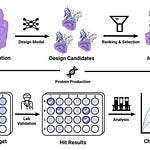
Venomics is a new deep learning–powered framework developed by Cesar de la Fuente and colleagues at the University of Pennsylvania to discover antimicrobial peptides encrypted within venom proteins. By leveraging the APEX model, the team computationally mined over 16,000 venom proteins to generate more than 40 million venom-encrypted peptides (VEPs), identifying potent candidates for tackling multidrug-resistant bacteria.
This platform marks a breakthrough in combining molecular biodiversity and machine learning to uncover entirely new families of antibiotics with high activity and low toxicity.
Key Innovations and Capabilities
1. Sequence-to-Function Discovery from Venom Proteomes
Predict: APEX, a deep neural model, predicts strain-specific antimicrobial activity (MIC) for millions of short venom-derived peptides.
Filter: The model identifies candidates based on low sequence similarity to known antimicrobial peptides (AMPs) while retaining desirable charge and hydrophobic profiles.
Validate: Top VEPs were synthesized and tested experimentally across multiple resistant bacterial strains and in a live infection mouse model.
2. Exploring New Sequence and Property Space
Over 40 million peptides were generated by slicing 16,123 venom proteins from cone snails, spiders, snakes, and scorpions.
Unsupervised UMAP projections revealed that VEPs mapped to novel regions of sequence space, distinct from known AMPs.
ISOB- and UniProt-derived peptides were the most divergent and promising in terms of antimicrobial novelty.
3. Structurally Responsive Peptides
VEPs exhibited environment-sensitive secondary structures, transitioning from disordered forms to α-helices in membrane-mimicking conditions.
This helical folding in SDS and TFE environments aligns with classical AMP behavior, suggesting membrane-disruptive action.
Compared to peptides from previous APEX runs, these venom-derived candidates displayed greater conformational plasticity.
4. Mechanism of Action and In Vivo Efficacy
Most VEPs acted by depolarizing bacterial membranes, a mechanism that bacteria find harder to evolve resistance against.
91% of tested VEPs showed activity against at least one clinical strain, including E. coli, P. aeruginosa, and MRSA.
Three lead peptides reduced bacterial load in a murine skin abscess model of Acinetobacter baumannii infection, matching the efficacy of polymyxin B.
5. Safety and Selectivity
Cytotoxicity and hemolysis assays demonstrated that several top-performing VEPs maintained high antimicrobial potency with minimal toxicity to human cells and red blood cells.
Venoms from spiders and cone snails contributed especially promising scaffolds with favorable selectivity indices.
Applications in Antimicrobial Drug Discovery
Rapid Screening: Venomics identifies membrane-active peptides without experimental screening, reducing time from data to discovery.
New Antibiotic Classes: Peptides with novel sequences and modes of action offer alternatives to traditional enzyme-targeting antibiotics.
Template Design: VEPs can serve as tunable scaffolds for future drug optimization, offering balance between efficacy and safety.
Mechanistic Probing: Predictive folding and membrane interaction profiles inform structure-function relationships directly from sequence.
Performance and Validation
Of 58 synthesized peptides, 53 were active against drug-resistant strains.
In vivo testing confirmed efficacy in a live infection model without adverse effects.
Compared to previous AMP discovery pipelines, Venomics delivered candidates with greater novelty, tunability, and structural insight.
Limitations and Future Directions
Current focus: Discovery from known venom proteomes using strain-specific MIC predictions.
Challenges: Interpretability of deep learning predictions remains limited, and some VEPs may have off-target effects via ion channel modulation.
Next steps: Incorporate large language models, expand training to negative examples, and pursue electrophysiological profiling for systemic safety.
Goal: Transform venom peptides into next-generation antibiotics using AI-driven screening, optimization, and design.
Why It Matters
The antibiotic pipeline has slowed, even as resistance rises. Venomics AI brings together the evolutionary power of animal toxins and the scalability of machine learning to discover antibiotics from natural sources previously too complex to analyze. With its ability to mine vast molecular libraries and surface novel, tunable candidates, Venomics represents a new frontier in computational antibiotic discovery.